Click the green plus () icon next to the Gene sets section header on the library file management page. Alternatively, click the Add library file button and choose Gene set from the Library type drop-down list.
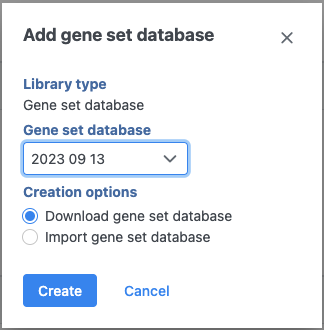
If you are using an assembly supported by Partek (e.g. human), a gene set from geneontology.org will appear in the Add gene set drop-down list (Figure 1). Select the Download gene set radio button and click Create.
Figure 1. For many model organisms, automatic downloads of gene sets are available from the Partek repository (source: http://geneontology.org/)
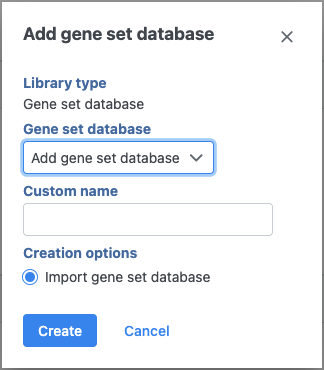
Figure 2. Adding a custom gene set
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
39 | rates |
Overview
Content Tools