Note that this task is for adding indexes for alignment to the whole genome. If you want to align to the transcriptome or another set of genomic features, see Adding Aligner Indexes Based on an Annotation Model.
- Bowtie
- Bowtie colorspace
- Bowtie 2
- TMAP
- BWA
- SHRiMP 2
- SHRiMP 2 colorspace
- Issac 2
- STAR
- STAR 2.4 1d
- GSNAP
- GSNAP v8
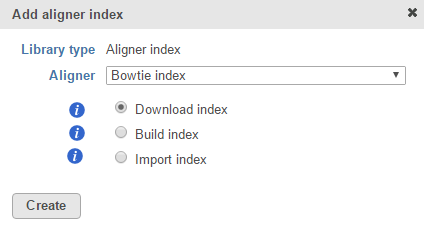
If you are using an assembly supported by Partek (e.g. human), there are three radio button options: Download index; Build index or Import index (Figure 1). Certain aligner indexes may not be available for automatic download because the file sizes are too large to download efficiently.
If available, select Download index and click Create to get the chosen reference aligner index from the Partek repository.
Alternatively, select Build index and click Create to build the reference aligner index. To build an aligner index, a reference sequence file must already be associated with the assembly. Depending on the aligner, you may have to specify further parameters. Consult the user documentation for each aligner for guidance (usually available on-line).
Alternatively, select Import index and click Create to add an aligner index from another source. An aligner index can be added from the Partek Flow Server, My Computer or a URL download link. The behavior of each option is similar to when importing a reference sequence (see Adding a Reference Sequence). When browsing for files on the Partek Flow server, only the files with relevant file extensions will be visible. This will vary for each aligner.
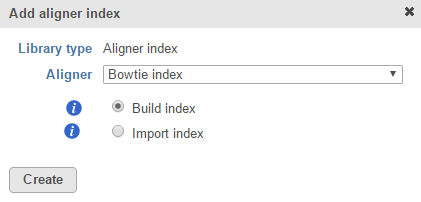
For custom assemblies (e.g. for non-model organisms), only the Build index and Import index options are available (Figure 2).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
40 | rates |