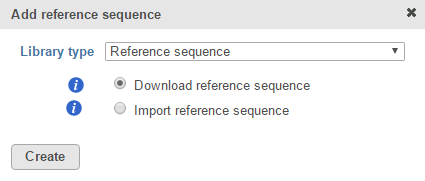
Click the green plus () icon next to the Reference Files section header and choose Reference sequence from the Library type drop-down list in the Add reference sequence dialog (Figure 1). Alternatively, click the Add library file button and choose Reference sequence from the Library type drop-down list. If a cytoband file is already associated with an assembly, Reference sequence will be the only option available in the Add reference sequence dialog and will not appear in a drop-down list (Figure 1).
If you are using an assembly supported by Partek (e.g. human), there are two radio button options: Download reference sequence and Import reference sequence (Figure 1). Select Download reference sequence and click Create to get the reference sequence from the Partek repository. Alternatively, select Import reference sequence and click Create to add the reference sequence from another source.
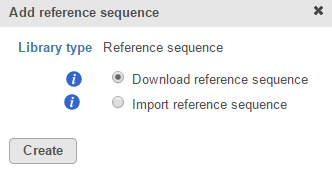
If you are using a custom assembly (e.g. for a non-model organism), only the Import reference sequence radio button will be available (Figure 2).
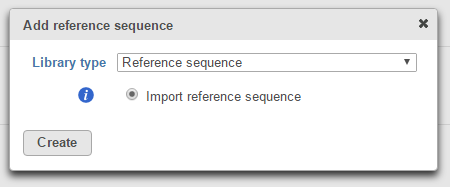
The Import reference sequence option allows you to add a reference sequence from the Partek Flow Server, My Computer or a URL download link (Figure 3).
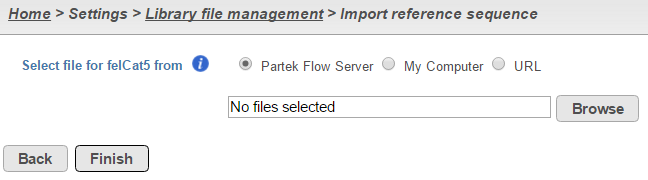
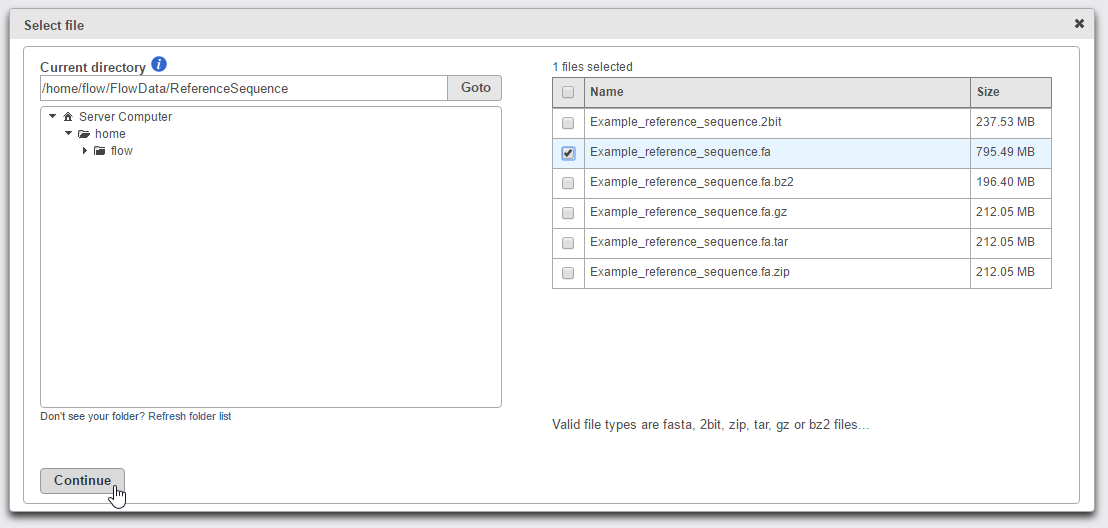
To add a reference sequence from the Partek Flow server, select Partek Flow Server, click Browse and navigate to the directory storing the reference sequence file(s) (Figure 4). Only files with the correct file extensions (.fa, .2bit or any compressed format) will be displayed in the file browser. Select the checkbox next to the file name(s) you wish to import and click Continue (Figure 4). Click Finish to add the selected reference sequence file(s) (Figure 3).
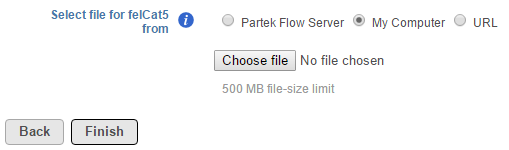
To upload a reference sequence from your local computer select My computer, click Choose file and navigate to the location of the file(s) on your local machine (Figure 5). It is not advisable to upload large files through the web browser, as this will likely result in an error (e.g. an upload timeout). Administrative users are able to set a limit on the upload file size via System preferences. The default file size limit is 500 MB. Please note that only one file can be uploaded at a time. Click Finish to add the selected reference sequence file(s) (Figure 5).
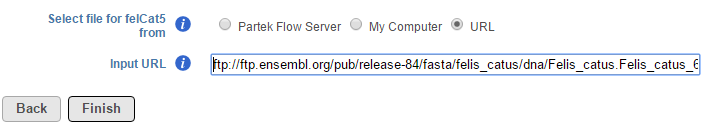
To download a reference sequence (e.g. from a public repository such as ENSEMBL), select URL and input the URL address (Figure 6). Make sure to include the full URL address, starting with the transfer protocol (e.g. http:// or ftp://) and ending with the file name. It is advisable to find the download link address, copy it to the clipboard and paste it into the Input URL box. Click Finish to add the selected reference sequence file(s) (Figure 6).
Note that you can specify multiple fasta files (e.g. one file per chromosome) and Partek Flow will concatenate them. If a fasta file is imported, a 2bit version of the file will automatically be created and vice-versa. Partek Flow will also accept a range of compressed file formats (gzip, bzip2, zip, tar). If a compressed file is imported, it will automatically be uncompressed and processed.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
42 | rates |