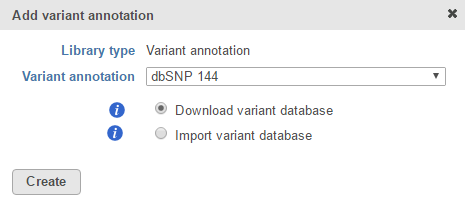
If you are using a human - hg19 assembly, variant annotation databases from various sources will appear in the Variant annotation drop-down list (Figure 1). Available variant annotation database sources include:
- dbSNP
- Kaviar
- NHLBI Variant Server
- 1000 Genomes
Multiple versions of the above databases are available. For human - hg38, only dbSNP is currently available. This list is periodically updated.
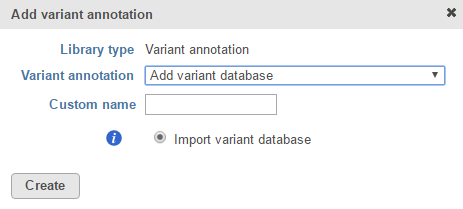
If you prefer to add a custom variant annotation database, perhaps from another source or 'gold-standard' validated variants, choose Add variant database from the Variant annotation drop-down list (Figure 2). Name the variant annotation database by typing into the Custom Name box and click Create. Characters such as $ * | \ : " < > ? / % cannot be used in custom names. A variant annotation database can be added from the Partek Flow Server, My Computer or a URL download link. The behavior of each option is similar to when importing a reference sequence (see Adding a Reference Sequence). When browsing for files on the Partek Flow server, only the files with relevant file extensions will be visible (.vcf and various compressed formats).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
41 | rates |