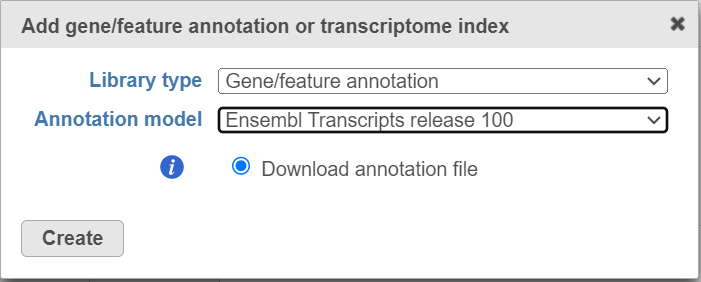
If you are using an assembly supported by Partek (e.g. human), annotation models from a variety of commonly used sources (e.g. ENSEMBL, GENCODE) will appear in the Annotation model drop-down list in the dialog. Choose an annotation model, select the Download annotation file radio button and click Create (Figure 1).
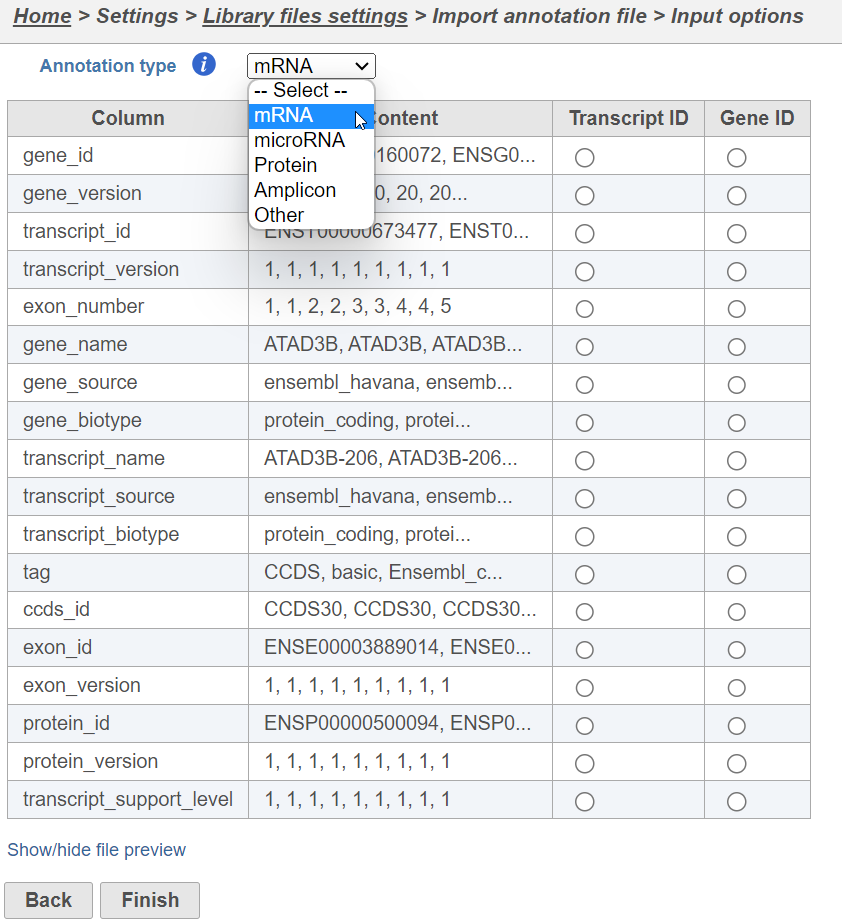
Select the source of annotation file and click Next. If the annotation file format is gtf, gff, gff3 or bed, a preview of the first 20 rows of the file will be shown on the screen (Figure 3). You must then specify the type of annotation file by choosing an option from the Annotation type drop-down list (Figure 3). Choose mRNA for whole transcriptome annotations, where both gene and transcript level information are present. Choose microRNA for precursor or mature microRNA transcript annotations. Choose Amplicon for targeted amplicon sequencing annotations. Choose Other for annotation files that do not fall into the other three categories (e.g. lncRNA).
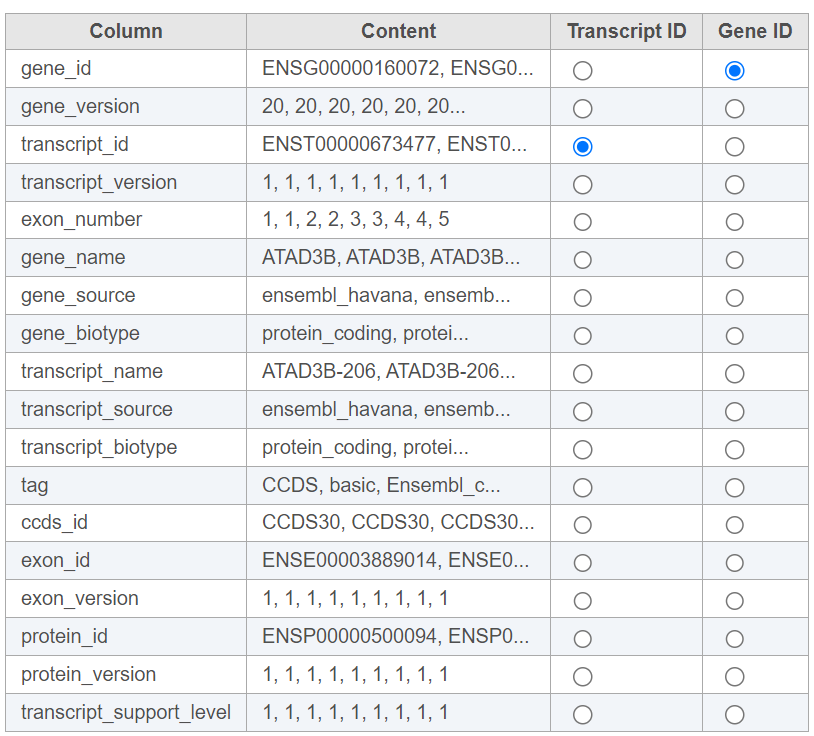
For text annotations with both gene- and transcript-level information select mRNA from the drop-down menu. You can manually specify which column corresponds to the Transcript ID or Gene ID by selecting the corresponding radio button next to the column with that information (Figure 4).
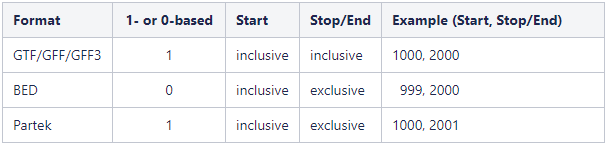
Genome coordinates for annotation models stored in Partek Flow are 1-based, start-inclusive, and stop-exclusive. This means that the first base position starts from one, the start coordinate for a feature is included in the feature and the stop/end coordinate is not included in the feature. These are the genome coordinates that are printed in various task reports and output files when an annotation model is involved in the task. When custom annotation files are added to Partek Flow, the genome coordinates are converted into this format. The coordinates are converted back if necessary for a specific task. Figure 5 shows how the genome coordinates vary between different annotation formats.
Figure 5. Comparison of genome coordinate formats. The example column shows the coordinates for a feature that starts at position 1000 (inclusive) and stops at position 2000 (inclusive).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
41 | rates |