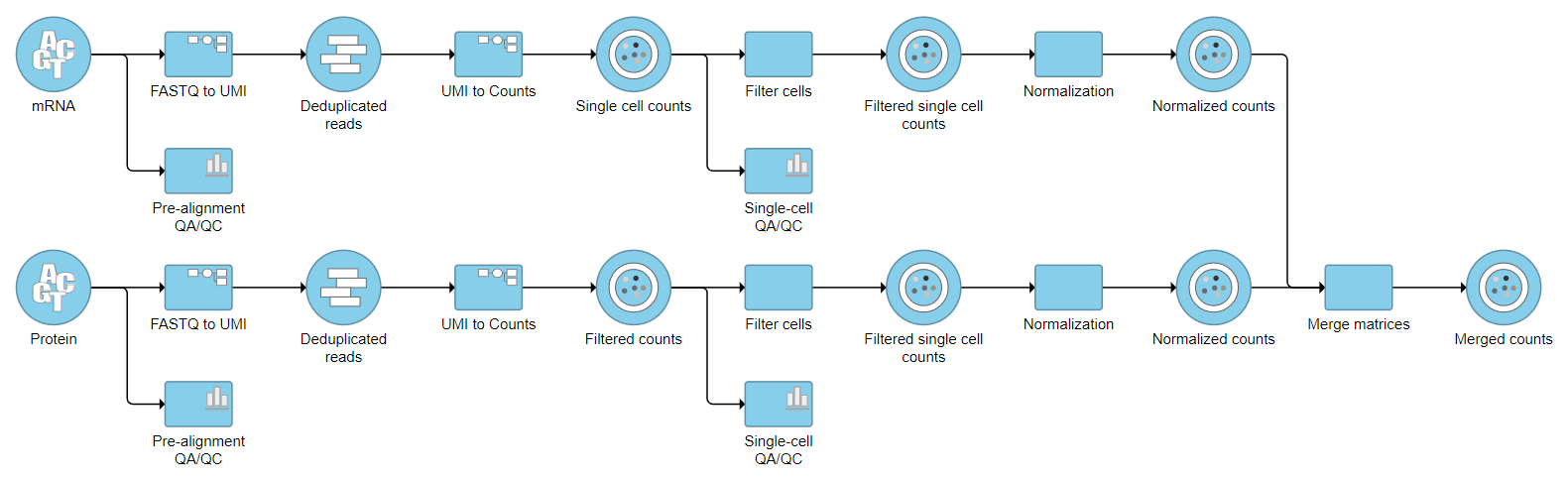
In projects with multiple data types for each sample such as CITE-Seq, Merge matrices can be used to combine two data nodes with different data types into one data node.
Prerequisites for Merge matrices
- There must samples with more than one type of data in the project
- There must be separate counts data nodes for each data type
Running Merge matrices
- Click the first counts data node
- Click the Pre-analysis tools section of the toolbox
- Click Merge matrices
A second data node of a different type than the first selected data node is chosen automatically. The second data node can selected manually using the Select data node button.
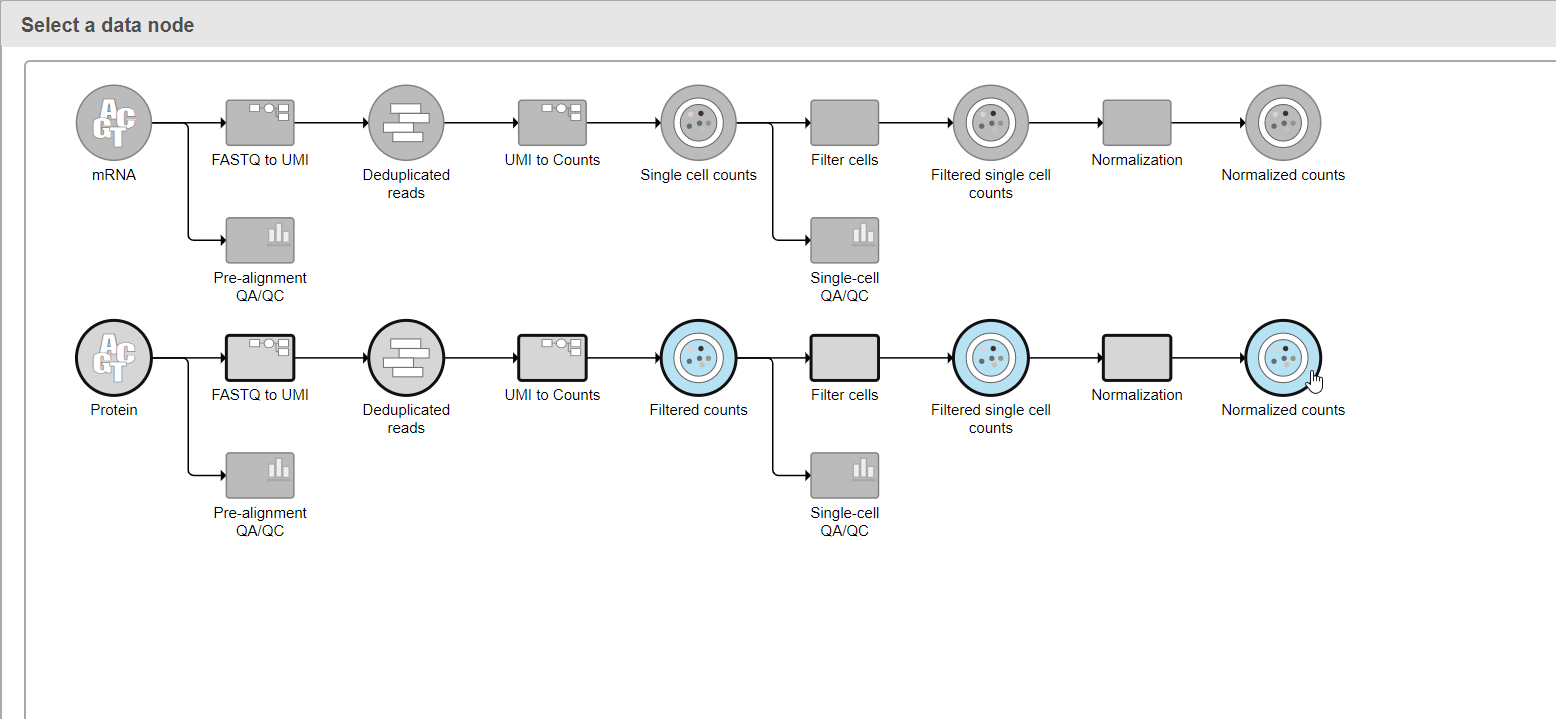
- Click Select data node to choose the second data node you want to merge (Figure 1)
- Double-click the second data node to select it (Figure 2)
- Click Finish to run
A Merged counts data node is produced (Figure 3).
Once two data types have been merged, they can be split using Split matrix.
For a practical example using Merge matrices, please see our tutorial on Analyzing CITE-Seq Data,
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
9 | rates |