In complex projects, different data matrices (e.g. observations on rows and features on columns) need to be merged in order to achieve the analysis goals. For example, two cell populations were identified on separate branches of the analysis pipeline and to combine them before any joint downstream steps, the expression matrices have to be combined. Alternatively, two assays (gene expression and protein expression) were performed on the same cells so the expression matrices have to be merged for joint analysis.
Merge matrices task is located in the Pre-analysis tools section of the toolbox and it can handle two scenarios: Merge cells/samples and Merge features (Figure 1). To start, select the first data node on the pipeline (e.g. single cell counts) and then select the Merge matrices task.
Merge Cells/Samples
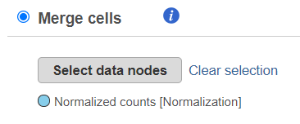
To use the Merge cells option, the data matrices (one or more) that are to be merged with the currently selected one should have the same features (e.g. genes), but distinct cells. Push the Select data nodes button and Partek Flow will display a preview of the pipeline; the data nodes that can be merged are shown in color of the branch, other data nodes are disabled (greyed out). Left click on the data node that you want to merge with the current one and push the Select button, you can select multiple data nodes to merge. The selected node(s) will be shown under the Select data nodes button (Figure 2). If you made a mistake, use the Clear selection icon. Push Finish to proceed.
Merge Features
To use the Merge features option, the data matrices (one or more) that are to be merged with the currently selected one should have the same cells (or samples), but distinct features (e.g. gene and protein expression). Push the Select data nodes button and Partek Flow will display you a preview of the pipeline; the data nodes that can be merged are shown in color of the branch, others are disabled (greyed out). Left lick on the data node that you want to merge with the current one and push the Select button. The selected node will be shown under the Select data nodes button. Repeat the procedure if you would like to merge additional nodes. If you made a mistake, use the Clear selection icon. Push Finish to proceed.
Task Output
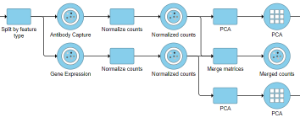
The output of the Merge matrices task is a Merged counts data node (Figure 3).
For a practical example using Merge matrices, please see our tutorial on Analyzing CITE-Seq Data.
Alternative paths
Depending on your goal, you may want to consider a different approach. For example, data matrices based on two different assays (e.g. gene and protein expression) can be combined using Find multimodal neighbors. Instead of merging two (or more) cell populations by using Merge cells, you may want to use filtering (Filtering > Filter groups) to filter out the populations that you do not consider relevant / filter in the populations of your interest.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
24 | rates |
1 Comment
Ivan Lukic
Good to go. Thanks, Paul!