In ChIP-seq or ATAC-seq analysis, after peak regions detected, quantify regions will report the read count from the peaks for each sample. The quantify regions algorithm is the same as Quantify to annotation model (Partek E/M), the annotation model is location of the peaks.
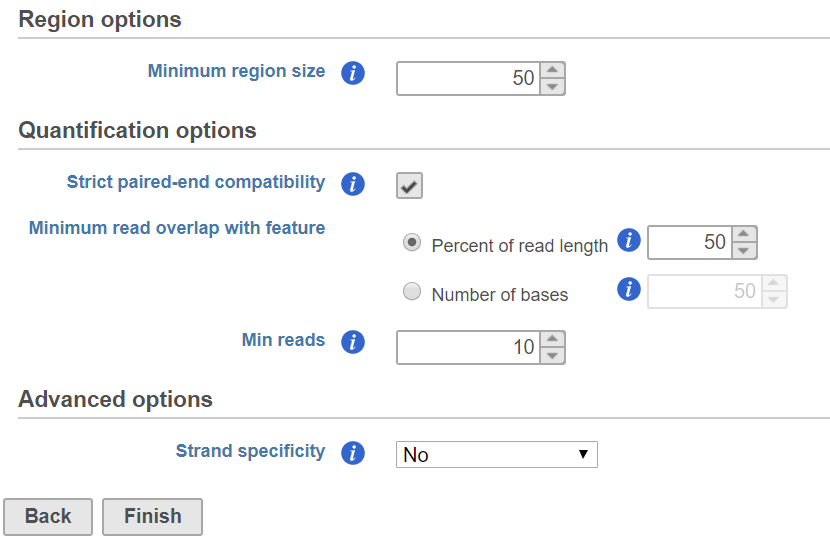
Quantify regions dialog
Click on peak data node, choose Quantify regions task in Quantification section. (Figure 1). In peak detection task, it report a set of peak regions in each sample individually. This task merged all the peaksets across all the samples to create a unique region list as a .bed file, in other words, it joins the break points of the regions across all the samples, each break point is either a start or stop location if a region. For instance. in sample A, a region detected on chromosome 1 from 100bp to 200bp, noted as chr1:100-200; in sample B, a region detected as chr1:105-210, to merge all the regions will get the following list of regions:
chr1:100-105 (region detected in sample A only)
chr1:105-200 (region detected in both sample A and sample B)
chr1:200-210 (region detected in sample B only)
When combine the regions from different samples, the regions are over segmented, some regions are very short. In the dialog, short regions can be filtered out by specify minimum region size, if a region is smaller than the specified cutoff, the region is removed from report.
Quantification options are the same as Quantify to annotation model (Partek E/M) dialog.
References
- Xing Y, Yu T, Wu YN, Roy M, Kim J, Lee C. An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res. 2006; 34(10):3150-60.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
1 | rates |