miRNAs regulate gene expression at the post-transcriptional level by base-pairing with the three prime untranslated region (3’ UTR) of the target gene, causing cleavage/degradation of the cognate mRNA or by preventing translation initiation. Integration of miRNA expression with gene expression data to study the overall network of gene regulation is vital to understanding miRNA function in a given sample. Partek Genomics Suite provides a platform that can analyze miRNA and gene expression data independently, yet allows data to be integrated for downstream analysis.This integrative analysis can be accomplished at several different levels. If you only have miRNA data, then Partek Genomics Suite can search the predicted gene targets in a miRNA-mRNA database like TargetScan to provide a list of genes that might be regulated by the differentially expressed miRNAs. Conversely, if you have only gene expression data, Partek Genomics Suite can use the same database to identify the microRNAs that putatively regulate those differentially expressed genes in a statistically significant manner. If you have gene expression data and miRNA data from comparable tissue/species, Partek Genomics Suite can combine the results of these separate experiments into one spreadsheet. Lastly, if the miRNA and mRNA from the same source was analyzed (as in this tutorial), then you may statistically correlate the results of miRNA and gene expression assays.
Finding putative genes regulated by miRNAs
This application is useful in the case where you have miRNA expression data, but not gene expression data. Using a database like TargetScan, microCosm, or a custom database, you can identify the list of genes that are predicted to be regulated by these differentially expressed miRNAs and then perform Biological Interpretation tasks on the list of genes.
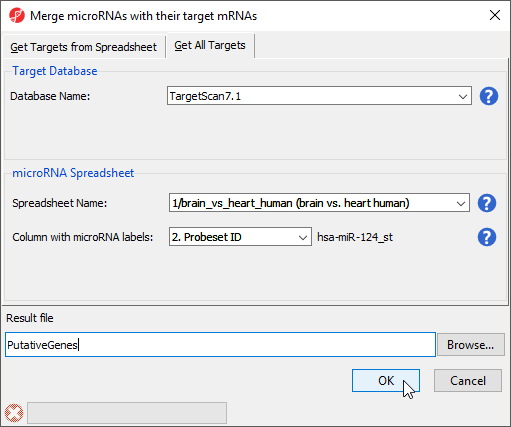
- Select Combine miRNAs with their mRNA targets from the miRNA Integration section of the miRNA Expression workflow
- Select the Get All Targets tab
- Select TargetScan7.1 for Database Name
- Select brain vs. heart human for Spreadsheet Name
- Set Column with microRNA labels to 2. Probeset ID
- Name the Result file PutativeGenes
- Select OK (Figure 1)
This will create a new spreadsheet PutativeGenes that contains a miRNA and a putative gene target in each row. Because each miRNA can regulate multiple genes, the list will be much longer than the input miRNA list. Because each row contains a gene, this spreadsheet can be analyzed using GO Enrichment and Pathway Enrichment tasks from the Biological Interpretation section of the workflow.
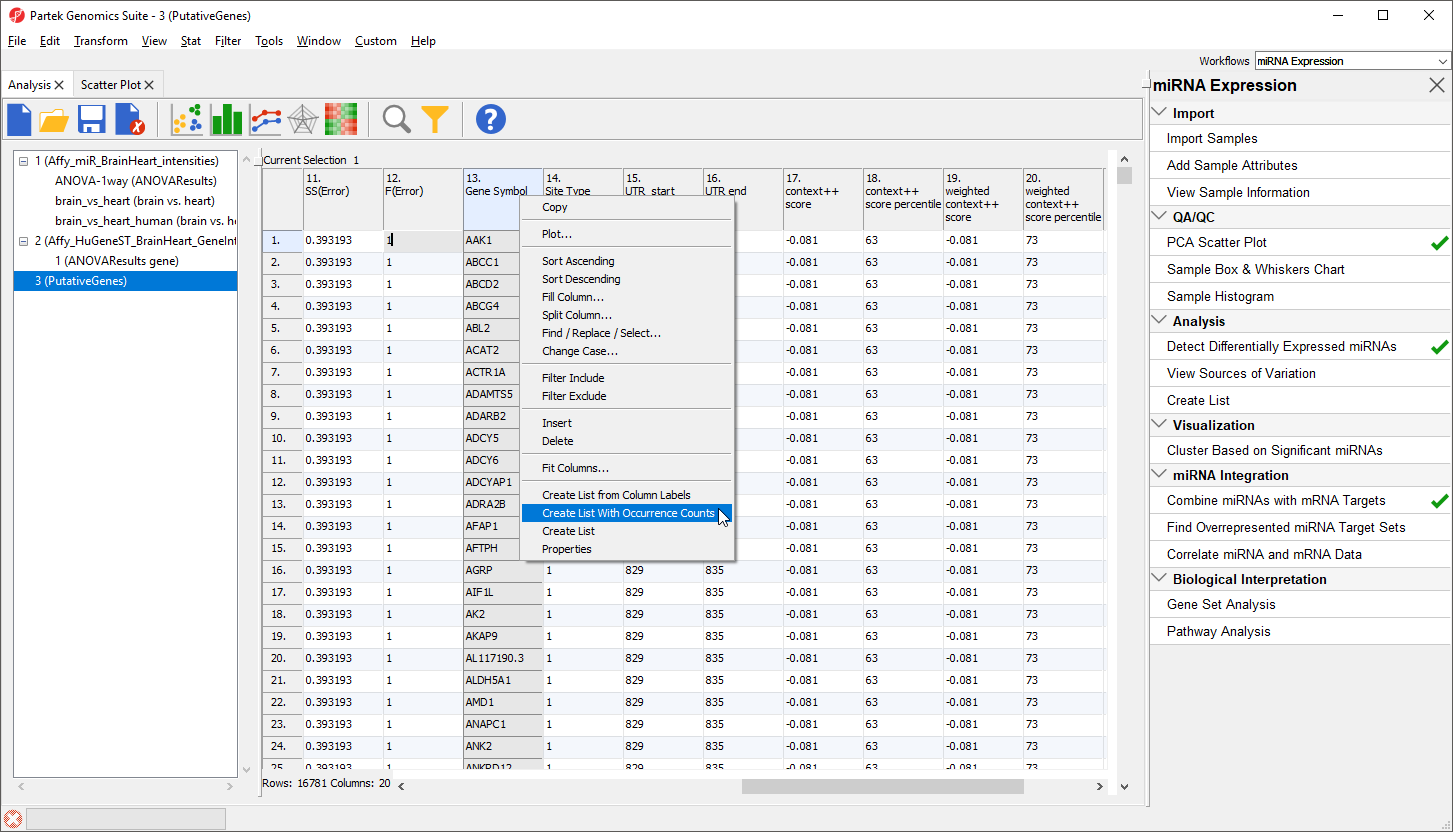
Another useful way to analyze this list is to determine which genes could be targeted by multiple miRNAs in the list. To do this:
- Right-click on the column 13. Gene Symbol header
- Select Create List With Occurrence Counts from the pop-up menu (Figure 2)
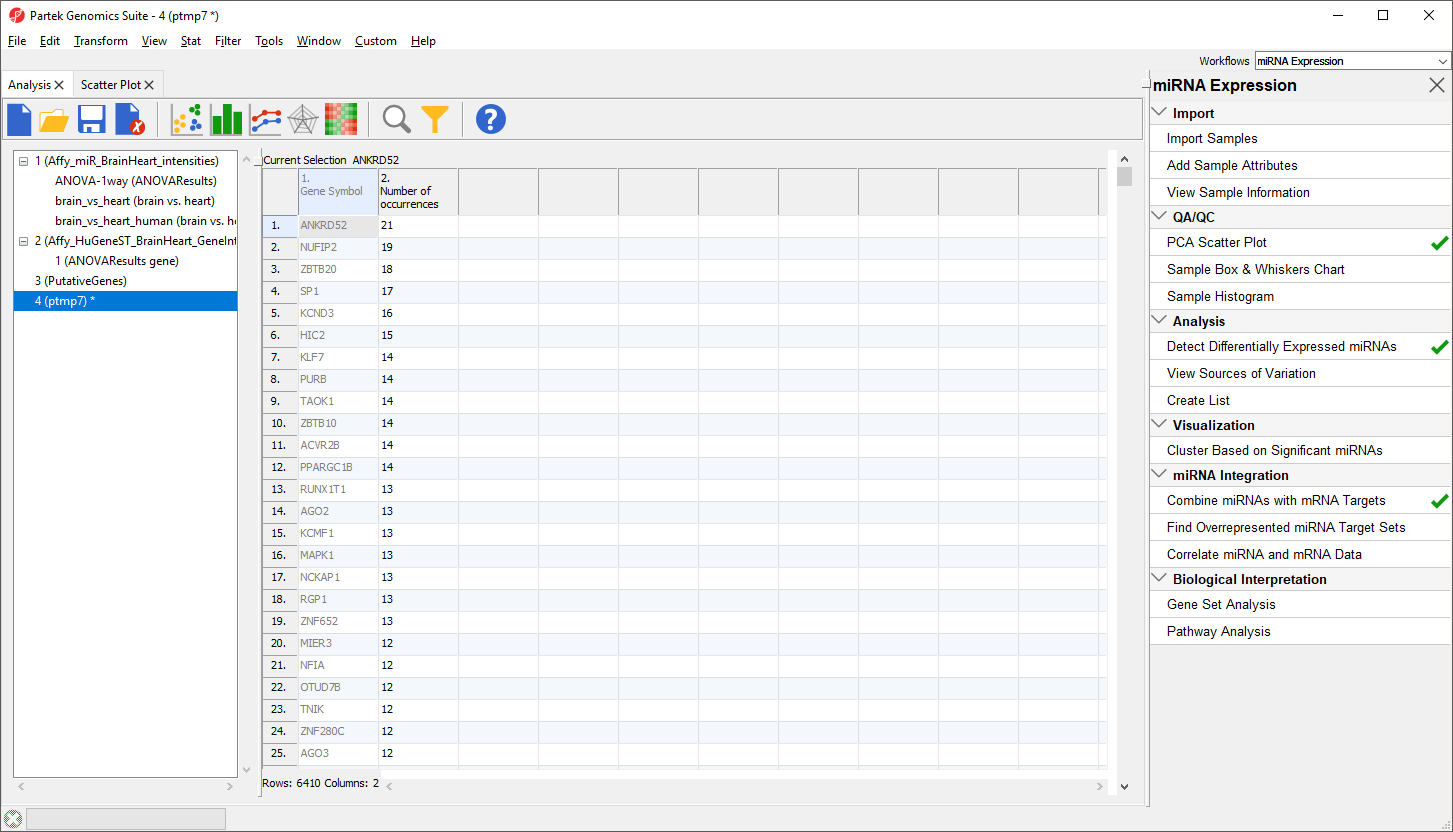
The new spreadsheet is a temporary spreadsheet listing each gene in alphabetical order and giving the occurance count of each. Sorting by descending order will list the gene with the most occurances first (Figure 3).
We will not be using this temporary spreadsheet moving forward. You can close the spreadsheet by selecting
Finding overrepresented miRNA targets sets from gene expression data
This application is useful when you only have gene expression results or a gene list of interest and are interested in identifying which miRNAs might regulated the genes. Using a databse like TargetScan, you can create a list of miRNAs that are statistically predicted to regulated those genes. miRNAs of particular interest could then be explored using a lower-throughput technique like RT-qPCR.
Using the gene list as input, a Fisher's Exact right-tailed p-value is calculated to show the overrepresentation of genes of interest for each miRNA in the database. THe smaller the p-value, the more overrepresented the miRNAs are for the dataset. Target associations are taken from a database, TargetScan in this example. If the input list is a filtered list of gnees from an ANOVA calculation, the parent spreadsheet is used to identify the background list of genes from the array. Gene in the array, but not in the significant gene list will be treated as background in the calculations.
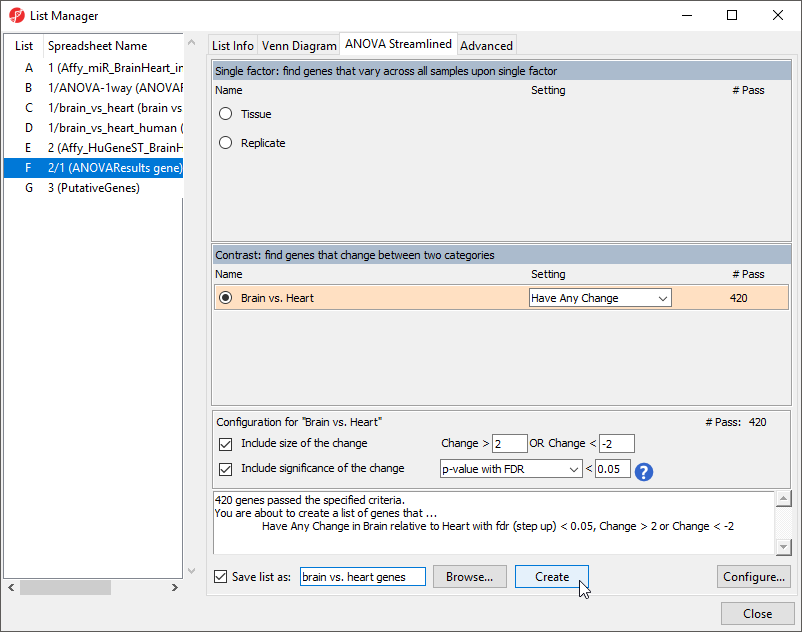
To begin, we need to create a list of significant genes using the ANOVAResults gene spreadsheet.
- Select the ANOVAResults gene spreadsheet in the spreadsheet tree
- Select Create List from the workflow
- Select Brain vs. Heart
- Set the Save list as to brain vs. heart genes
- Leave other fields at their default values (Figure 4)
- Select Create
- Select Close to exit the List Manager dialog
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |