Page History
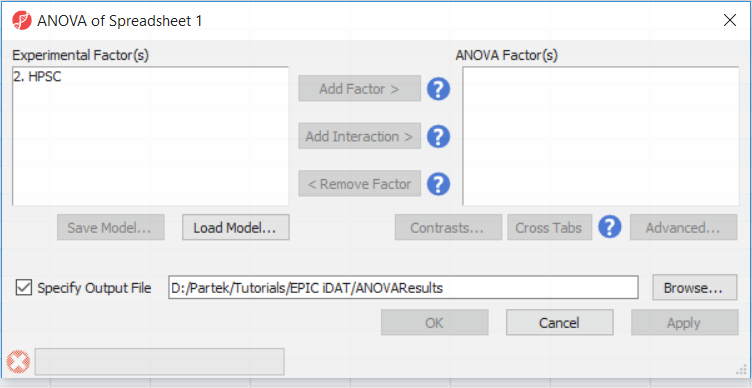
To detect differential methylation between CpG loci in different HSCP populations, go to Analysis > Detect Differential Methylation. In the ANOVA dialog (Figure 1), select Add Factor to move the factor 2. HPSC from Experimental Factor(s) to the ANOVA Factor(s) box.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Section Heading
Section headings should use level 2 heading, while the content of the section should use paragraph (which is the default). You can choose the style in the first dropdown in toolbar.
...
Overview
Content Tools