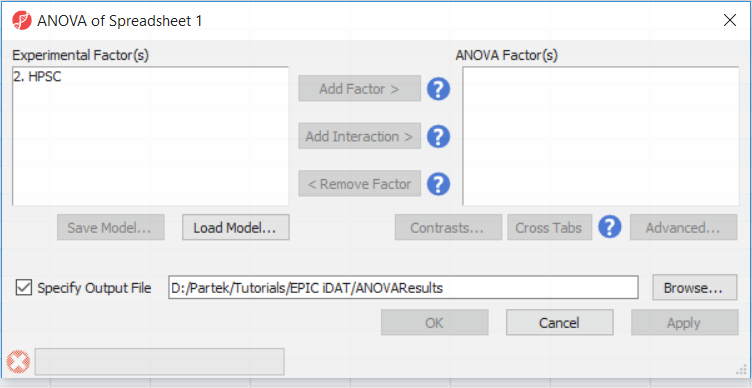
To detect differential methylation between CpG loci in different HSCP populations, go to Analysis > Detect Differential Methylation. In the ANOVA dialog (Figure 1), select Add Factor to move the factor 2. HPSC from Experimental Factor(s) to the ANOVA Factor(s) box.
Section Heading
Section headings should use level 2 heading, while the content of the section should use paragraph (which is the default). You can choose the style in the first dropdown in toolbar.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |