Page History
...
Click on peak data node, choose Quantify regions task in Quantification section. (Figure 1), you do not need to specify annotation model since the regions used to generate read counts is a list of peak locations from all the samples generated in the . In peak detection task.
If the bam file is imported, you need to select the assembly with which the reads were aligned to, and which annotation model file you will use to quantify from the drop-down menus (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
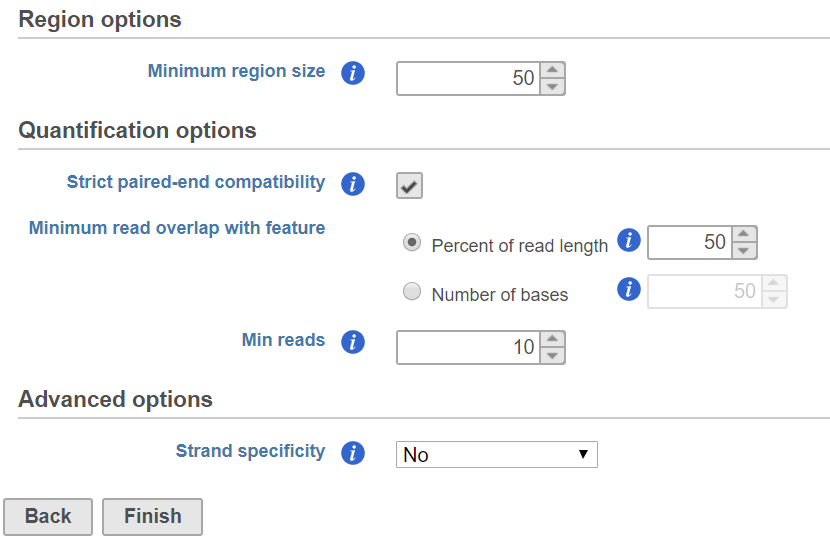
In the Quantification options section, when the Strict paired-end compatibility check button is selected, paired end reads will be considered compatible with a transcript only if both ends are compatible with the transcript. If it is not selected, reads with only one end have alignment that is compatible with the transcript will also be counted for the transcript .
If the Require junction reads to match introns check button is selected, only junction reads that overlap with exonic regions and match the skipped bases of an intron in the transcript will be included in the calculation. Otherwise, as long as the reads overlap within the exonic region, they will be counted. Detailed information about read compatibility can be found in the Understanding Reads tutorial.
Some library preparations reverse transcribe the mRNA into double stranded cDNA, thus losing strand information. In this case, the total transcript count will include all the reads that map to a transcript location. Others will preserve the strand information of the original transcript by only synthesizing the first strand cDNA. Thus, only the reads that have sense compatibility with the transcripts will be included in the calculation. We recommend verifying with the data source how the NGS library was prepared to ensure correct option selection.
In the options, forward means the strand of the read must be the same as the strand of the transcript while reverse means the read must be the complementary strand to the transcript (Figure 3). The options in the drop-down list will be different for paired-end and single-end data. For paired-end reads, the dash separates first- and second-in-pair, determined by the flag information of the read in the BAM file. Briefly, the paired-end Strand specificity options are:
- No: Reads will be included in the calculation as long as they map to exonic regions, regardless of the direction
- Auto-detect: The first 200,000 reads will be used to examine the strand compatibility with the transcripts. Two percentages are calculated: (1) the percentage of reads whose first-in-pair is the same strand as the transcript and second-in-pair is the opposite strand to transcript, (2) the percentage of reads whose first-in-pair is the opposite strand to transcript and second-in-pair is the same strand as the transcript. If the 1st percentage is higher than 75%, the Forward-Reverse option will be used. If the 2nd percentage is higher than 75%, the Reverse-Forward option will be used. If neither of the percentages exceed 75%, No option will be used
- Forward - Reverse: this option is equivalent to the --fr-secondstrand option in Cufflinks [1]. First-in-pair is the same strand as the transcript, second-in-pair is the opposite strand to the transcript
- Reverse - Forward: this option is equivalent to --fr-firststrand option in Cufflinks. First-in-pair is the opposite strand to the transcript, second-in-pair is the same strand as the transcript. The Illumina TruSeq Stranded library prep kit is an example of this configuration
- Forward - Forward: Both ends of the read are matching the strand of the transcript. Generally colorspace data generated from SOLiD technology would follow this format
The single-end Strand specificity options are:
- No: same as for paired-end reads
- Auto-detect: same as for paired-end reads. All single-end reads are treated as first-in-pair reads
- Forward: this option is equivalent to the --fr-secondstrand option in Cufflinks. The single-end reads are the same strand as the transcript
...
, it report a set of peak regions in each sample individually. This task merged all the peaksets across all the samples to create a unique region list as a .bed file, in other words, it joins the break points of the regions across all the samples, each break point is either a start or stop location if a region. For instance. in sample A, a region detected on chromosome 1 from 100bp to 200bp, noted as chr1:100-200; in sample B, a region detected as chr1:105-210, to merge all the regions will get the following list of regions:
chr1:100-105 (region detected in sample A only)
chr1:105-200 (region detected in both sample A and sample B)
chr1:200-210 (region detected in sample B only)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Minimum read overlap with feature can be specified in percentage of read length or number of bases. By default, a read has to be 100% within a feature. You can allow some overhanging bases outside the exonic region by modifying these parameters.
Min reads optioin is a filter, by default only the features whose sum of the reads across all samples that are greater than or equal to 10 will be reported. To report all the features in the annotation file, set the value to 0.
If the Report unexplained regions check button is selected, an additional report will be generated on the reads that are considered not compatible with any transcripts in the annotation provided. Based on the Min reads for unexplained region cutoff, the adjacent regions that meet the criteria are combined and region start and stop information will be reported.
In the annotation file, there might be multiple features in the same location, or one read might have multiple alignments, so the read count of a feature might not be an integer. Our white paper on the Partek E/M algorithm has more details on Partek’s implementation the E/M algorithm initially described by Xing et al. [1]
Quantify to annotation model (Partek E/M) output
Depending on the annotation file, the output could be one or two data nodes. If the annotation file only contains one level of information, e.g. miRNA annotation file, you will only get one output data node. On the other hand, if the annotation file contains gene level and transcript level information, such as those from the Ensembl database, both gene and transcript level data nodes will be generated. If two nodes are generated, the Task report will also contain two tabs, reporting quantification results from each node. Each report has two tables. The first one is a summary table displaying the coverage information for each sample quantified against the specified transcriptome annotation (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The second table contains feature distribution information on each sample and across all the samples, number of features in the annotation model is displayed on the table title (Figure 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The bar chart displaying the distribution of raw read counts is helpful in assessing the expression level distribution within each sample. The X-axis is the read count range, Y axis is the number of features within the range, each bar is a sample. Hovering your mouse over the bar displays the following information (Figure 6):
- Sample name
- Range of read counts, “[ “represent inclusive, “)” represent exclusive, e.g. [0,0] means 0 read counts; (0,10] means the range is greater than 0 count but less than and equal to 10 counts.
- Number of features within the read count range
- Percentage of the features within the read count range
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The coverage breakdown bar chart is a graphical representation of the reads summary table for each sample (Figure 7)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
In the box-whisker plot, each box is a sample on X-axis, the box represents 25th and 75th percentile, the whiskers represent 10th and 90th percentile, Y-axis represents the read counts, when you hover over each box, detailed sample information is displayed (Figure 8).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
...
- Sample name
- Range of read counts, “[ “represent inclusive, “)” represent exclusive
- Number of features within the read count range in the sample
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The box whisker and sample histogram plots are helpful for understanding the expression level distribution across samples. This may indicate that normalization between samples might be needed prior to downstream analysis. Note that all four visualizations are disabled for results with more than 30 samples.
The output data node contains raw reads of each sample on each feature (gene or transcript or miRNA etc.depends on the annotation used). When click on a output data node, e.g. transcript counts data node, choose Download data on the context sensitive menu on the right, the raw reads of transcripts can be downloaded in two different format (Figure 10):
Partek Genomics Suite project format: it is a zip file, do not manually unzip it, you can choose File>Import>Zipped project in Partek Genomics Suite to import the zip file into PGS.
Text file format: it is a .txt file, you can open the text file in any text editor or Microsoft Excel, each row is a transcript, each column is a sample.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| |||
When combine the regions from different samples, the regions are over segmented, some regions are very short. In the dialog, short regions can be filtered out by specify minimum region size, if a region is smaller than the specified cutoff, the region is removed from report.
Quantification options are the same as Quantify to annotation model (Partek E/M) dialog.
References
- Xing Y, Yu T, Wu YN, Roy M, Kim J, Lee C. An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res. 2006; 34(10):3150-60.
...