Before performing pathway enrichment, we need to create a gene list from the ANOVA results.
Creating a list of significant genes
- Select Gene Expression from the workflows drop-down menu
- Select the ANOVAResults gene spreadsheet
- Select Create Gene List from the Analysis section of the Gene Expression workflow
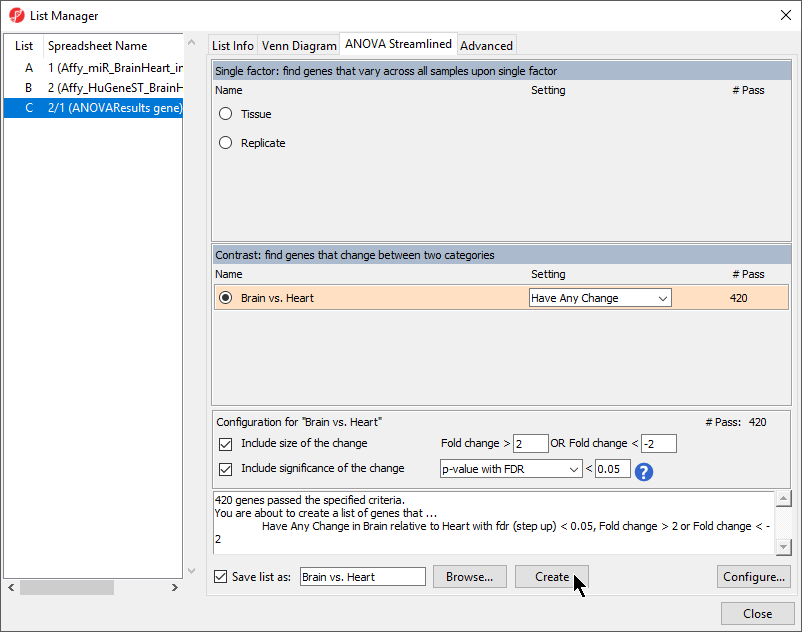
- Select Brain vs. Heart from the List Manager dialog (Figure 1) leaving the other options as defaults
- Select Create
A new list of 420 genes will be created as a child spreadsheet of 1 (ANOVAResults gene).
- Select Close to exit the List Manager dialog
Performing pathway enrichment analysis
- Select the new gene list, Brain vs. Heart
- Select Pathway Analysis from the Biological Interpretation section of the Gene Expression workflow
- Select Next > to continue with Pathway Enrichment
Pathway Enrichment is the only option available for a gene list. To learn more about the other option, Pathway ANOVA, see the Gene Ontology ANOVA tutorial, which follows the same procedure as Pathway ANOVA.
- Select Next > to continue with the Brain vs. Heart spreadsheet
- Select Next > to continue with default settings for Fisher's Exact test
- Select Next > to continue with Homo sapiens and 4. Gene Symbol as parameters
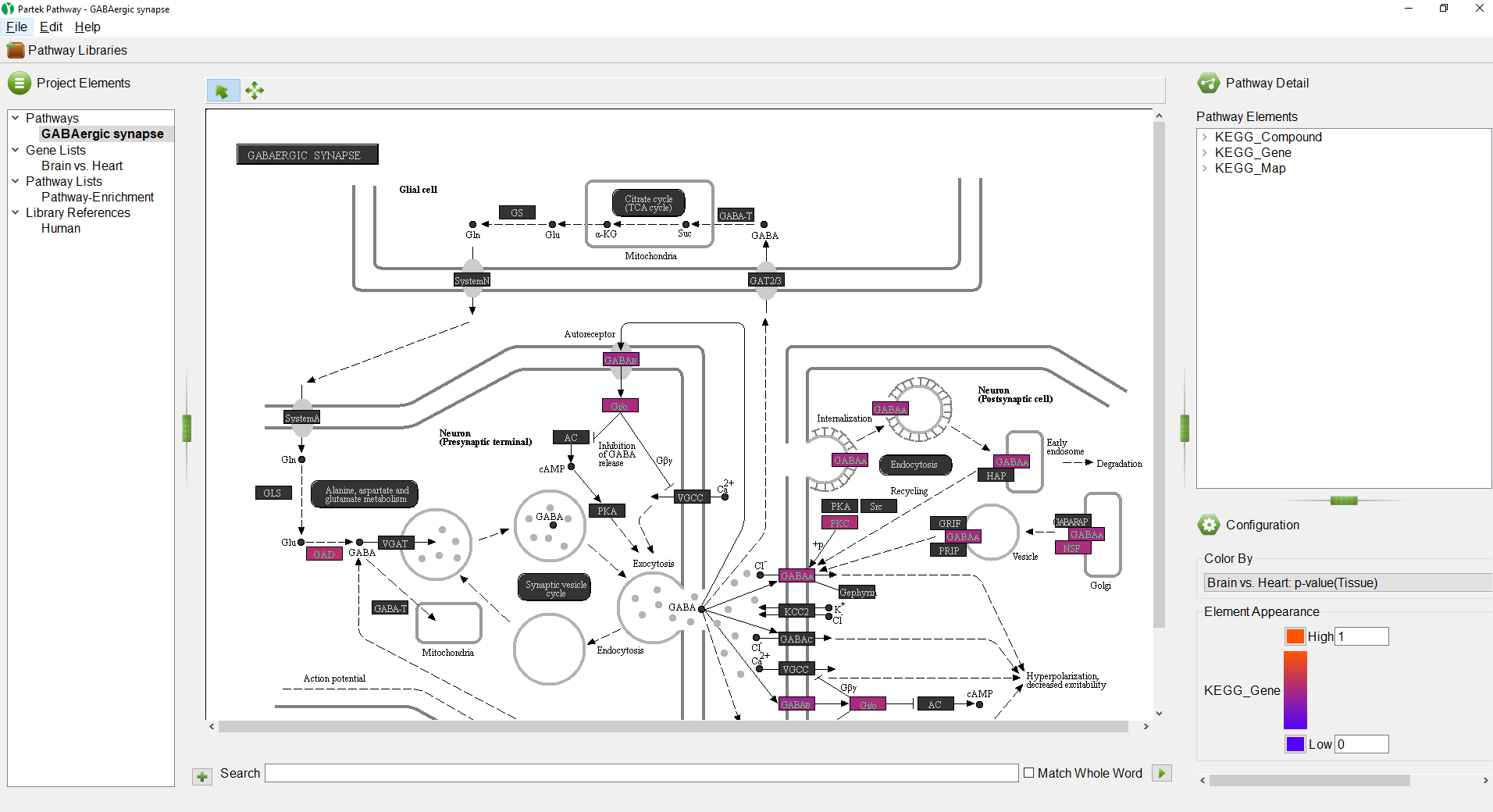
Partek Pathway will now open. If this is your first time using Partek Pathway on the selected species, Partek Pathway will automatically download the KEGG information needed for the analysis. Once the pathway enrichment calculation has been performed, a new spreadsheet, Pathway-Enrichment.txt, will be added as a child spreadsheet of Brain vs. Heart and Partek Pathway will launch (Figure 2).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
42 | rates |