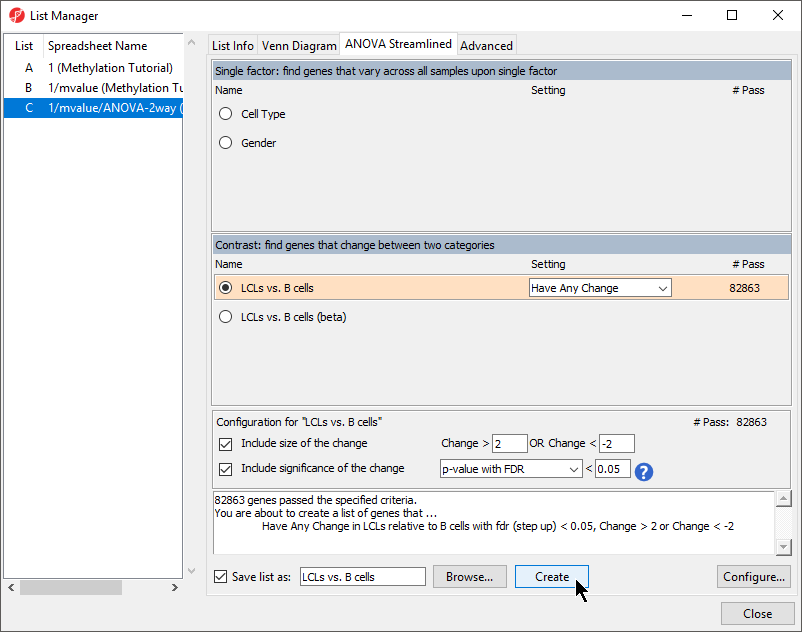
To analyze differences in methylation between our experimental groups, we need to create a list of deferentially methylated loci.
- Select Create Marker List from the Analysis section of the Illumina BeadArray Methylation workflow
- Select LCLs vs. B cells (Figure 1)
Figure 1. Creating a list of significantly differentially methylated loci
- Leave Include size of the change selected and set to Change > 2 OR Change < -2
- Leave Include significance of the change selected and set to p-value with FDR < 0.05
- Select Create
- Select Close to exit the list manager
The new spreadsheet LCLs vs. B cells (LCLs vs. B cells) will open in the Analysis tab.
It is best practice to occasionally save the project you are working on. Let's take the opportunity to do this now.
- Select File from the main command toolbar
- Select Save Project...
- Specify a name for the project, we chose Methylation Tutorial, using the Save File dialog
- Select Save to save the project
Saving the project saves the identity and child-parent relationships of all spreadsheets displayed in the spreadsheet tree. This allows us to open all relevant spreadsheets for our analysis by selecting the project file.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
34 | rates |
Overview
Content Tools