The Non-parametric ANOVA task uses the Kruskal-Wallis and Dunn's tests to identify differentially expressed genes between two groups. Note that such rank-based tests are generally advised for use with larger sample sizes.
Running the task
To invoke the Non-parametric ANOVA, select any count-based data nodes, these include:
- Gene counts
- Transcript counts
- Normalized counts
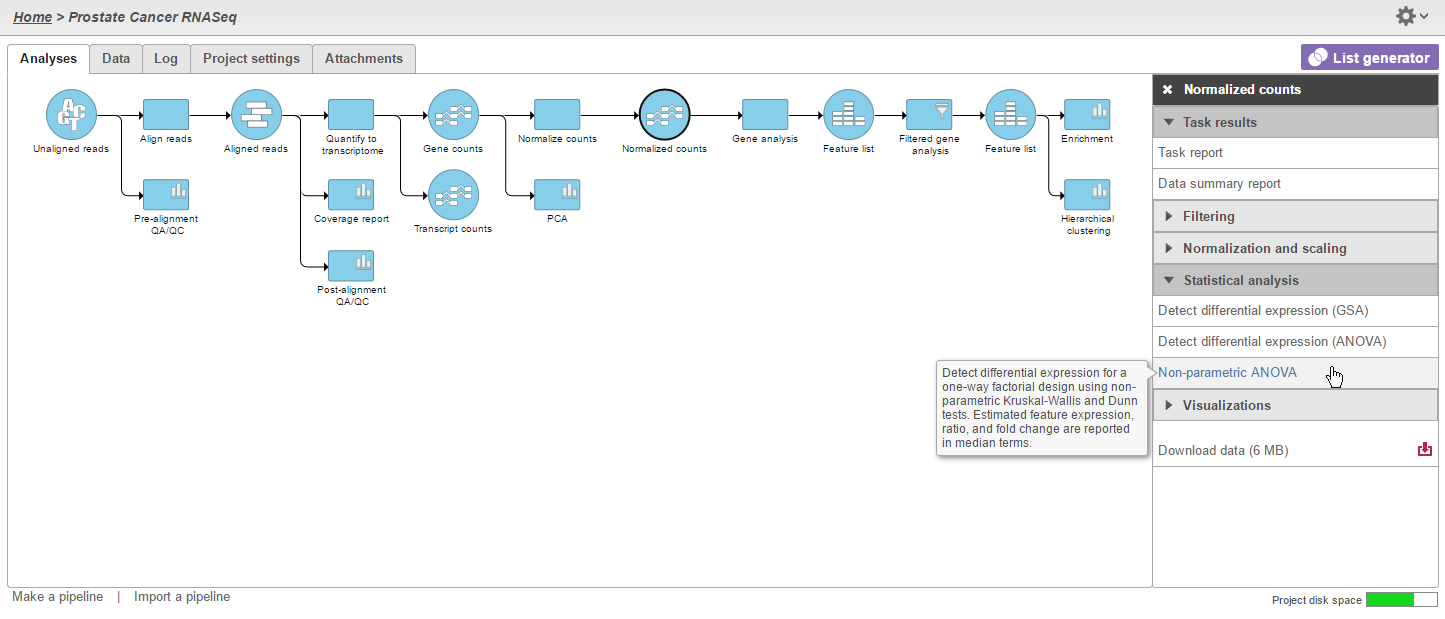
Select Non-parametric ANOVA under the Statistical analysis section of the context-sensitive menu (Figure 1).
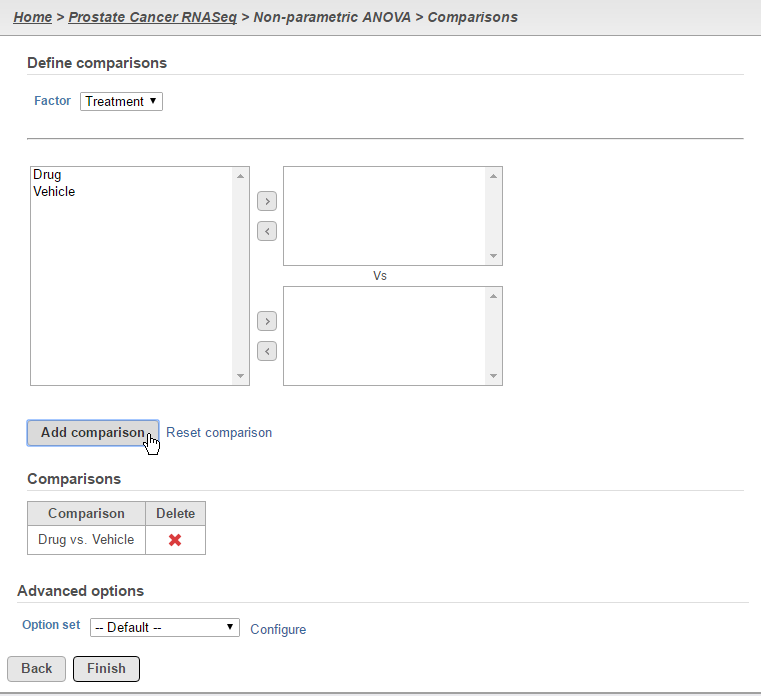
Select a specific factor for analysis and click the Next button (Figure 2). Note that this task can only take into account one factor at a time.
Define the desired comparisons between groups and click the Finish button (Figure 3). Note that comparisons can only be added between single group (i.e. one group per box).
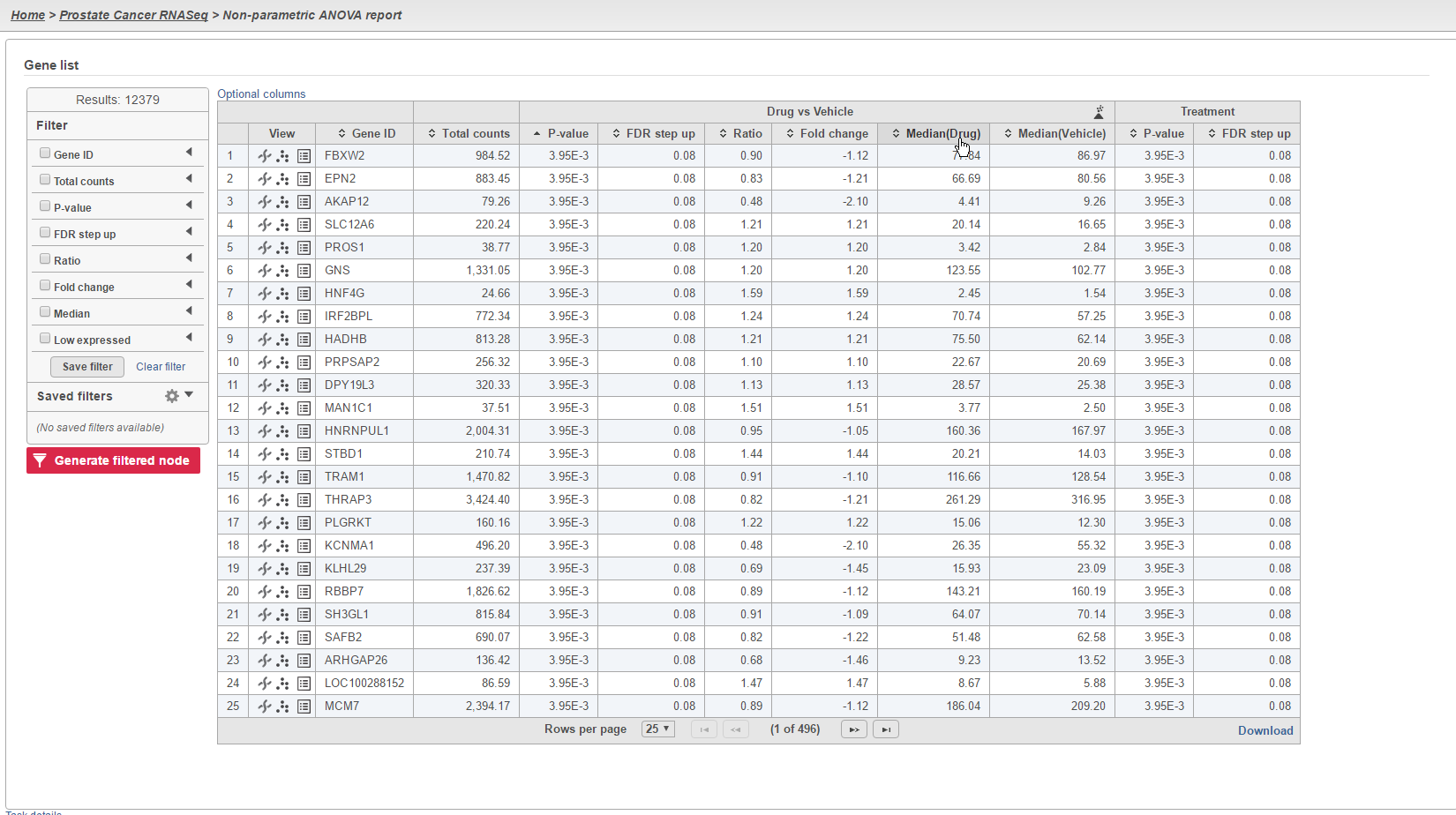
The results of the analysis will appear similar to other differential expression analysis results. However, the column to indicate mean expression levels for each group will display the median instead (Figure 4).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |