Partek Flow supports the import of filtered gene-barcode matrices generated by 10x Genomics' Cell Ranger pipeline. The following document will illustrate how to perform this import.
Below is a video summarizing the import of these files:
Importing matrices into Partek Flow (this Market Exchange Format is popular for public repositories)
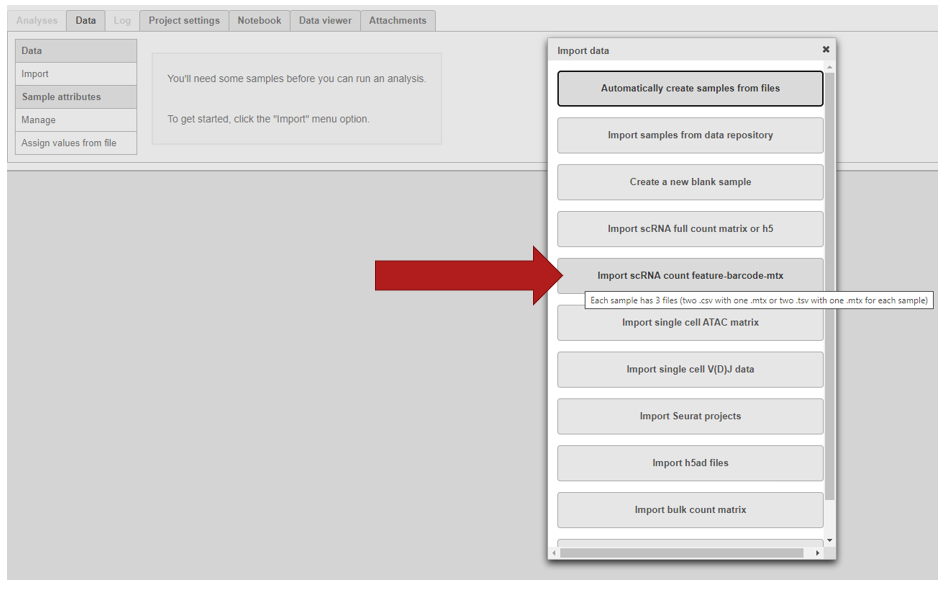
To import the matrices into Partek Flow, create a new project and click Import > Import scRNA count feature-barcode-mtx.
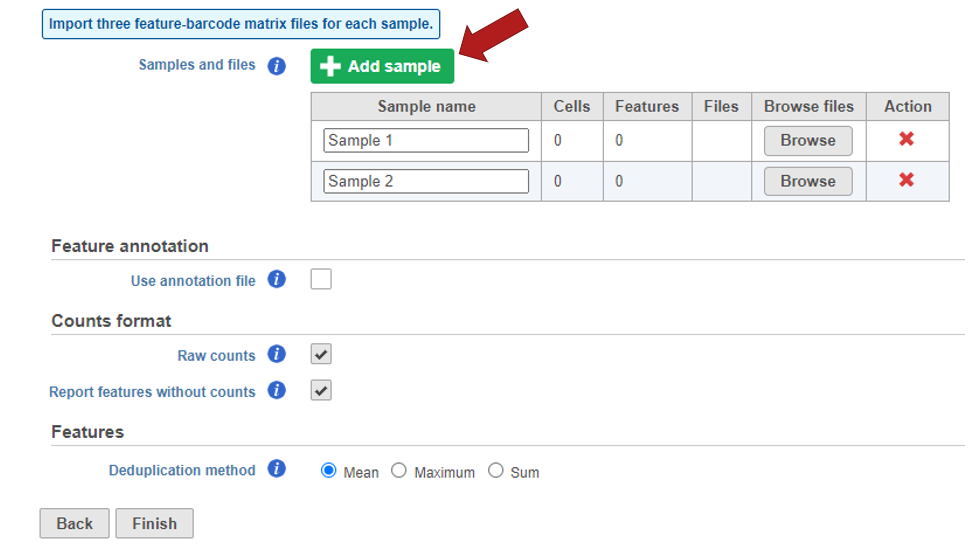
Samples can be added using the Add sample button. Each sample should be given a name and three files should be uploaded per sample using the Browse button.
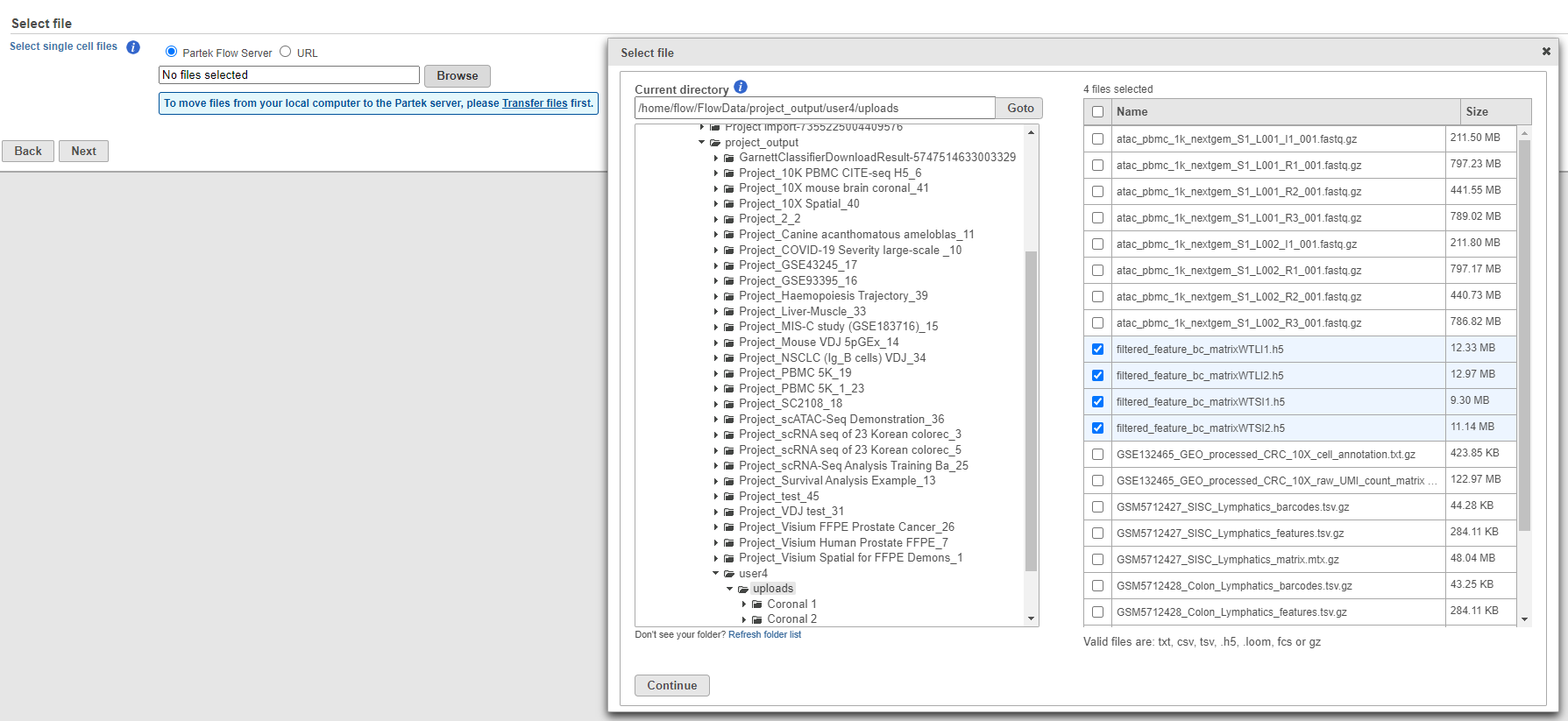
If you have not already, transfer the files to the server to be accessed when you click Browse. Use the Transfer files button on the Partek® Flow® homepage or use the transfer files link to upload data to the server. Navigate to the folder containing your filtered matrices. Use drag and drop or click My device to browse files from the device. Click Browse to modify the upload directory. The upload directory should be specified, known, and distinguishable for project file management. You will return to this directory and access the files to import them into a project.
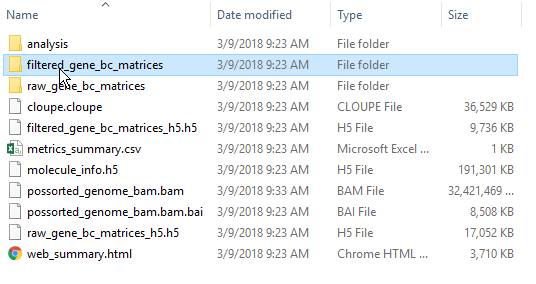
By default, the Cell Ranger pipeline output will have a folder called filtered_gene_bc_matrices (Figure 3). It is helpful to rename and organize the files prior to transfer using the File browser.
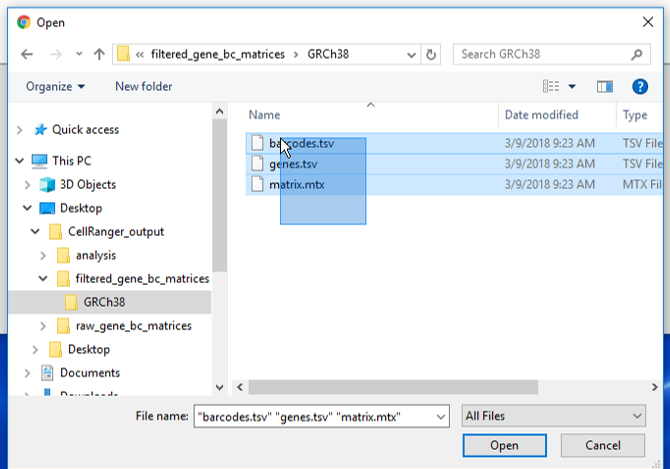
There are folders nested within the matrix folder, typically representing the reference genome it was aligned to. Navigate to the lowest subfolder, this should contain three files:
- barcodes.tsv
- genes.tsv
- matrix.mtx
Select all 3 files for import into Partek Flow
Specify the annotation file used when running the pipeline.
Click Finish when you have completed configuration. This will queue the import task.
Importing matrices in h5 format (this Hierarchical Data Format is recommended and ideal for multiple samples)
The Cell Ranger pipeline can also generate the same filtered gene barcode matrix in h5 format. This gives you the ability to select just one file per matrix and select multiple matrices to import in batch. To import an h5 matrix, select the Import scRNA full count matrix or h5 option. Browse for the files and modify any configuration options. Remember the files need to be transferred to the server.
This feature is also useful for importing multiple samples in batch. Simply put all h5 files from your experiment on a single folder, navigate to the folder and select all the matrices you would like to import.
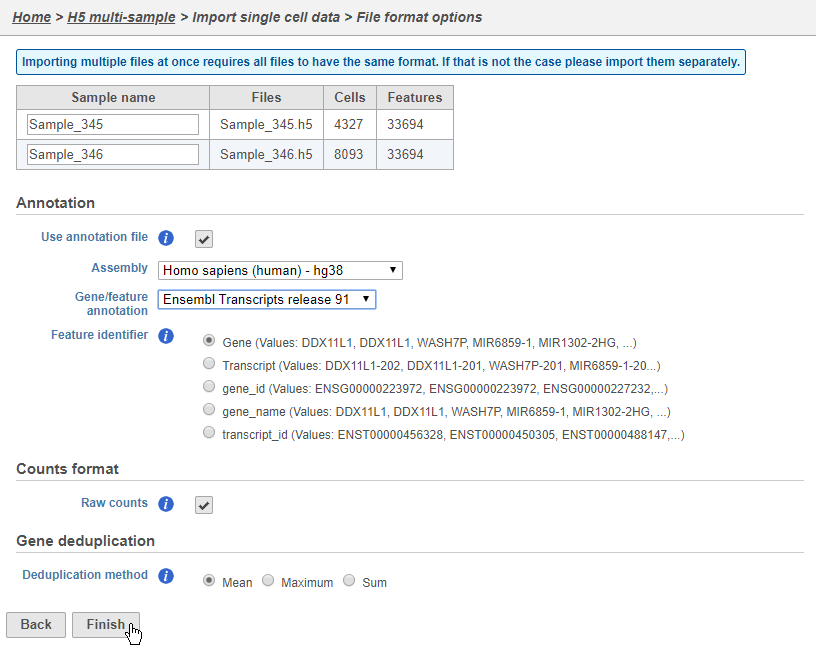
Configure all the relevant sample metadata, including sample name and the annotation that was used to generate the matrices, and click Finish when completed. Note that all matrices must have been generated using the same reference genome and annotation to be imported into the same project.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |