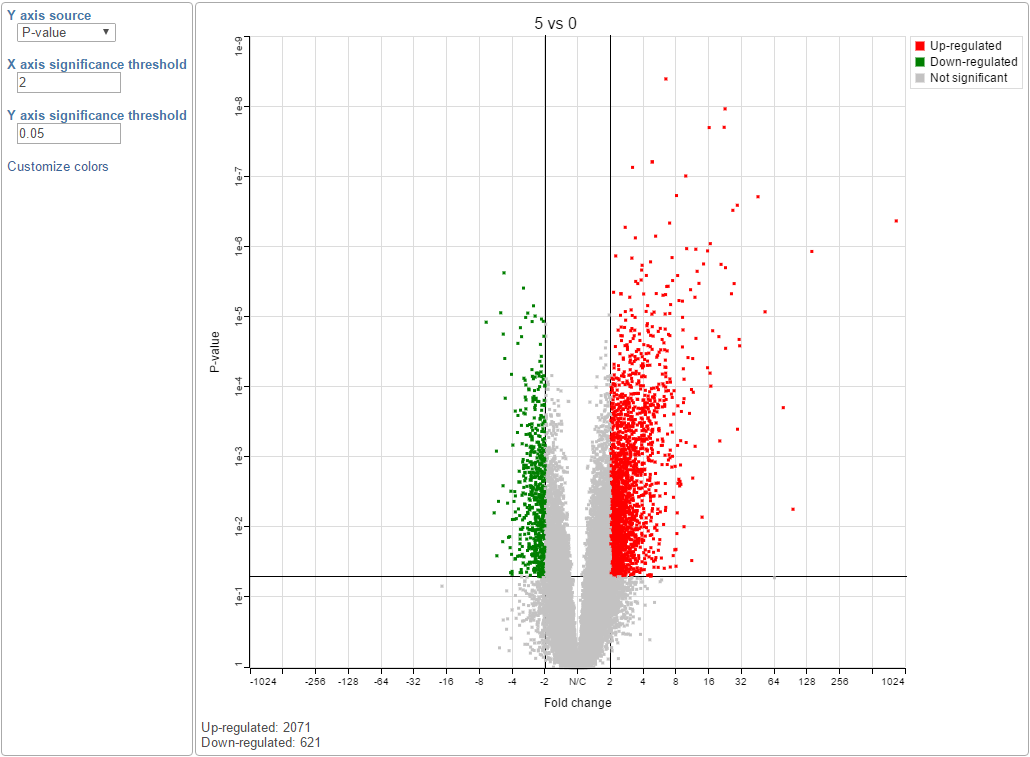
The volcano plot is a special 2-D scatter plot used to visualize significance and the magnitude of changes in features (e.g. genes or transcripts) within a given comparison. By convention, the X-axis represents the fold change between the two groups and is on a log2 scale. On the other hand, the Y-axis shows negative log10 of the p-values from the statistical test of the comparison.
As a prerequisite for invoking the volcano plot, you must run a Differential gene expression analysis. When setting-up the analysis, include all relevant contrasts you would like to view the volcano plots for.
Once the analysis is complete, open the resulting Feature list data node. Navigate to the Gene list section of the page and click on the volcano plot icon, located above the header of a Fold change column (Figure 1).
You can manually configure the Y axis source from the drop-down list and change the threshold of p-value and fold change, then the position of the lines and number of features in the significant areas will be updated accordingly. Click on the Customize colors can manually change the color of the dots (features) in the different areas.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |