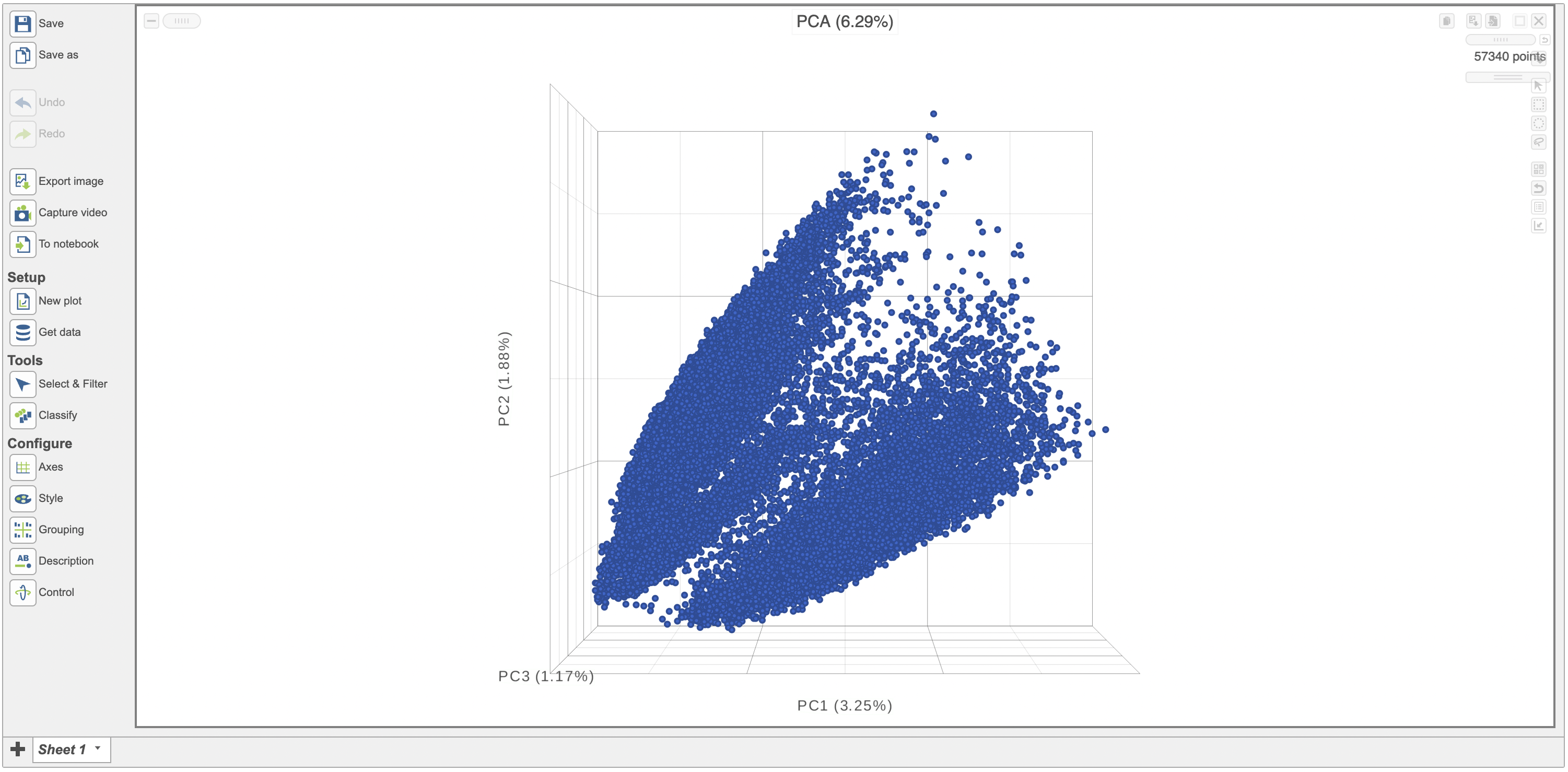
After performing exploratory analyses such as PCA, UMAP and t-SNE is is helpful to visualize the results on a scatterplot. This can help visually assess the source of variation affecting the results of an experiment, classify cells and select samples for downstream analysis. Here we have a PCA scatterplot generated from the analysis of 12 samples from a scRNA sequencing study.
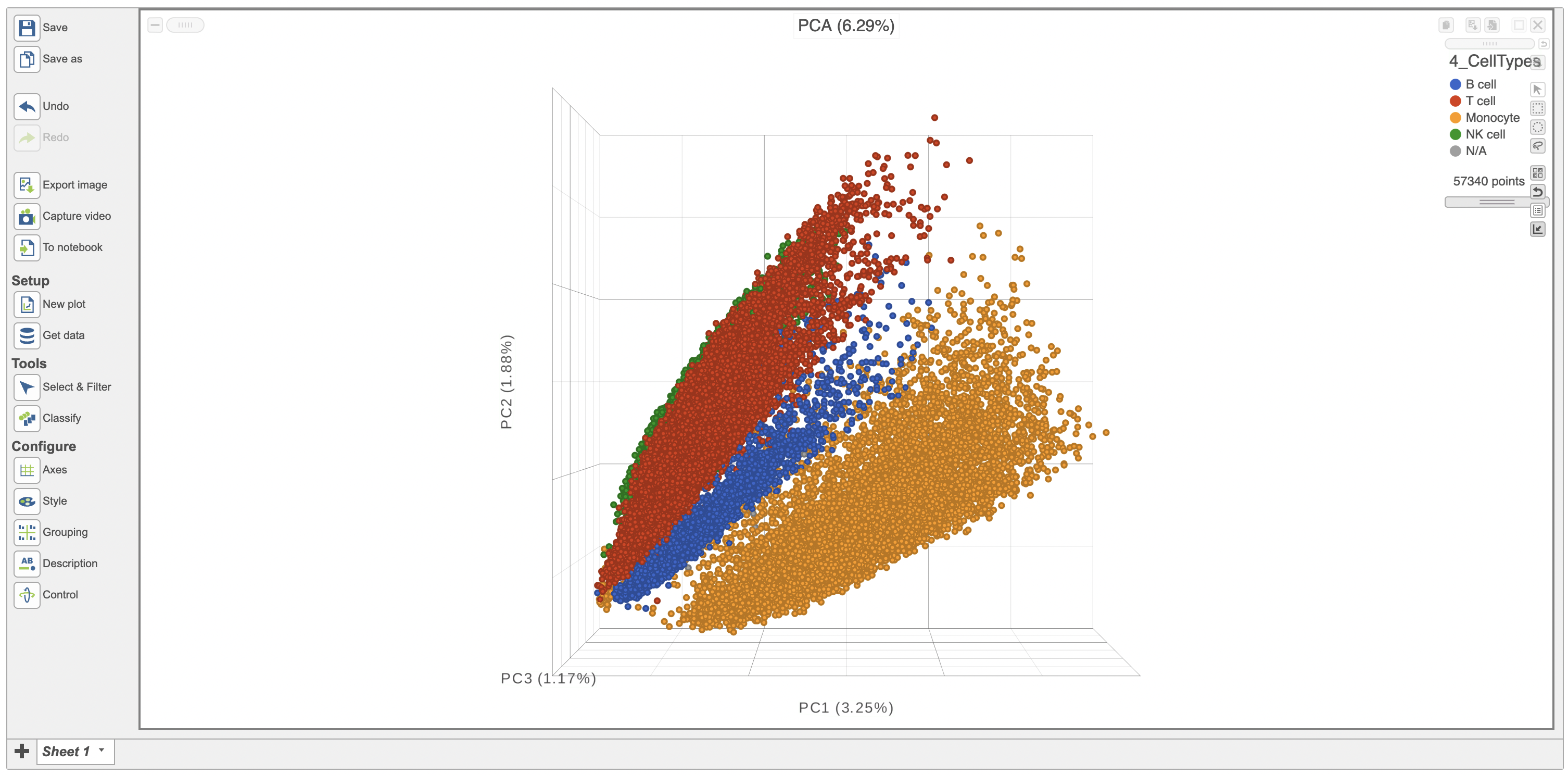
The Configure > Style menu on the left can then be used to color the features in the scatterplot based on an attribute (Figure 2). In this case, Figure 3 shows the cells being colored based on their cell-type.
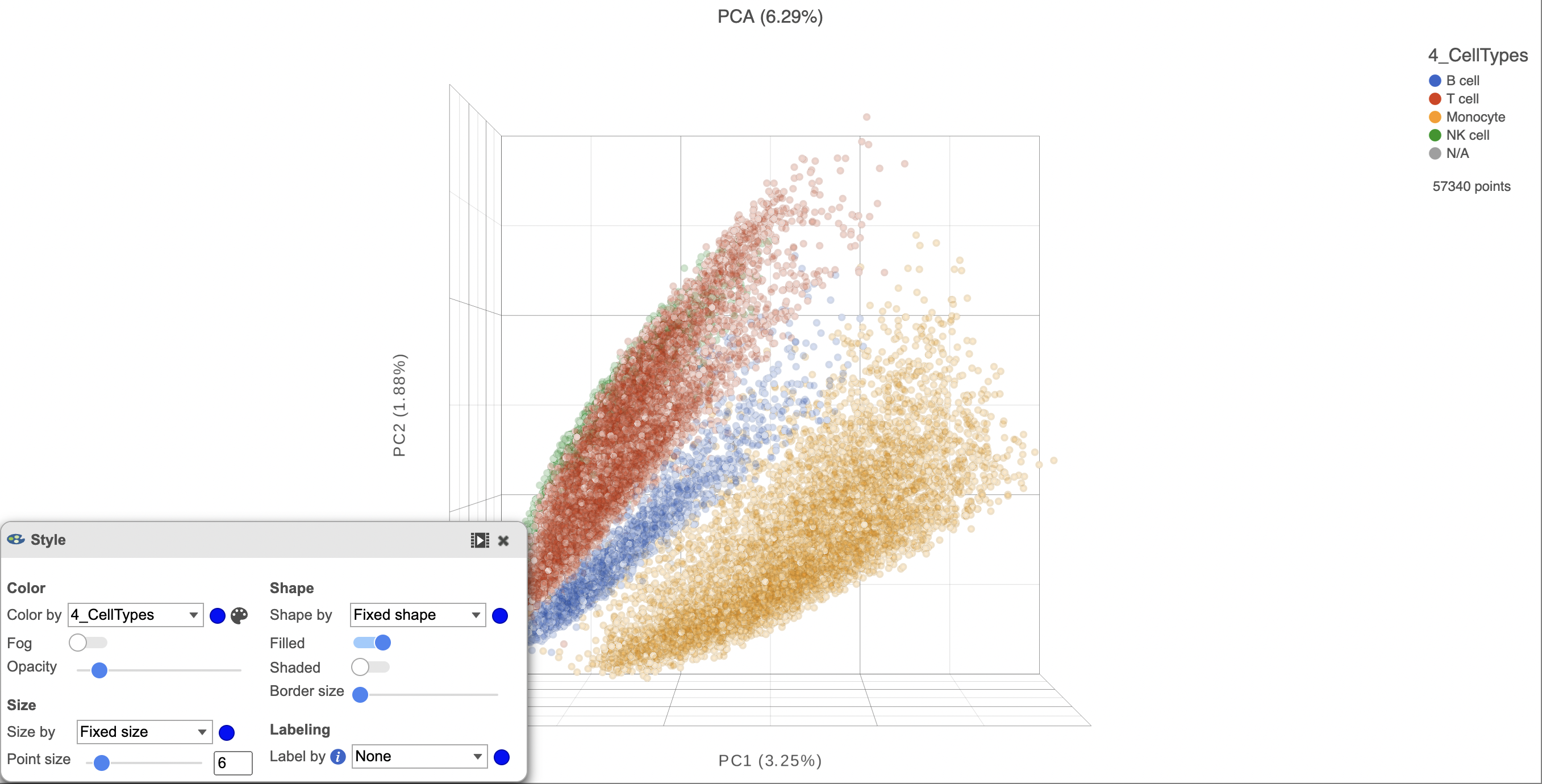
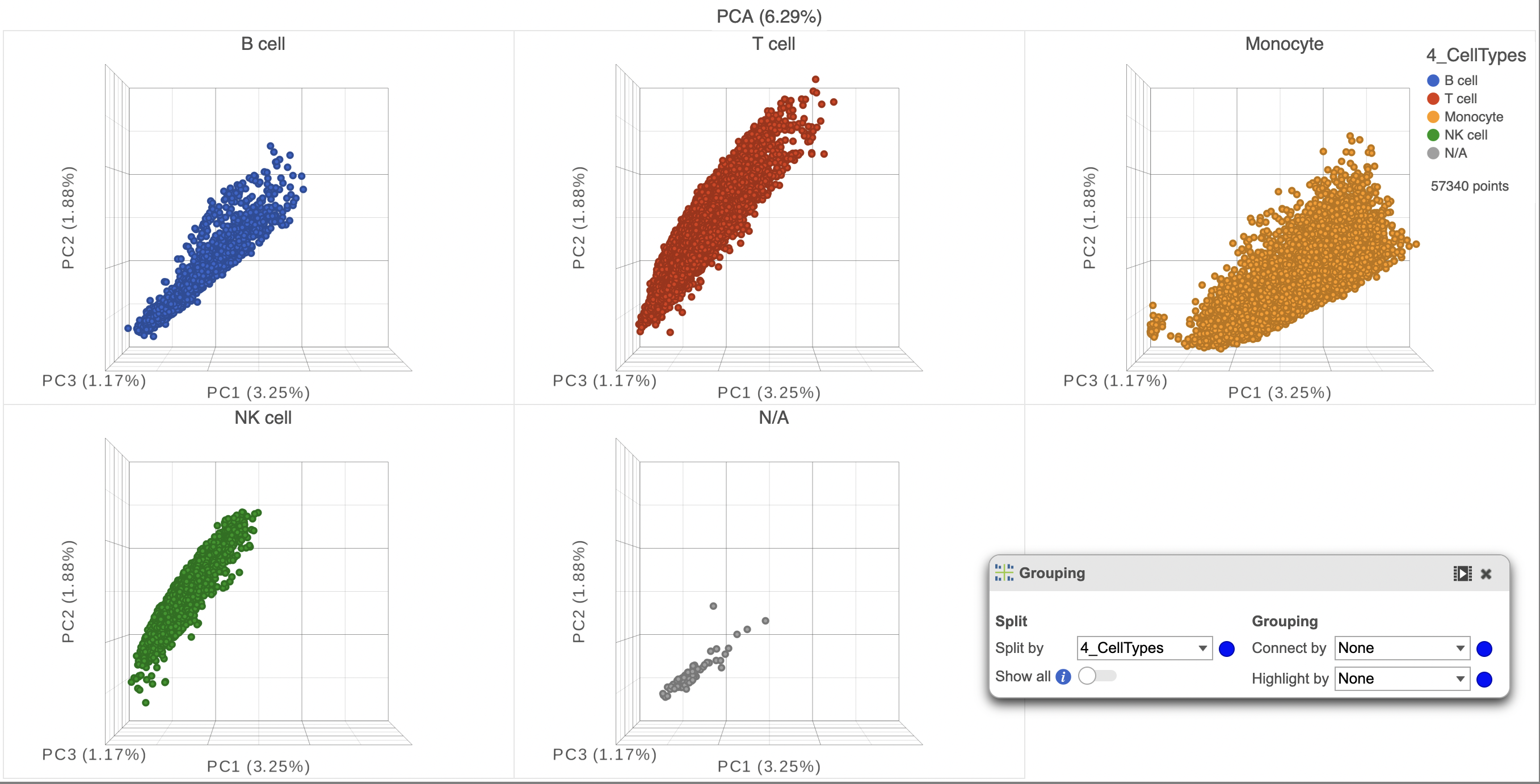
Additionally, you can adjust the opacity of the points to better assess the density across the groups (Figure 4). It is also possible to split the plot based on the same attribute in the Configure > Grouping menu (Figure 5)