Create a new Project
Let's start by creating a new project.
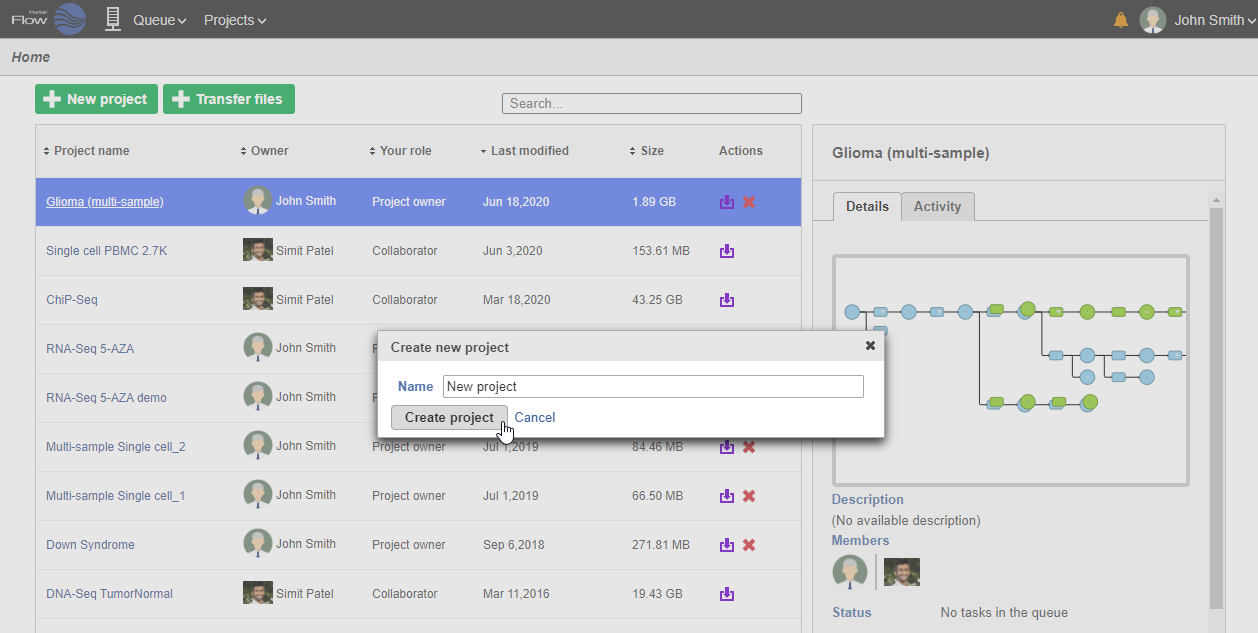
- On the Home page, click New project (Figure 1)
- Give the project a name
- Click Create project
Import data
- Under the Data tab, click Import data
- Click Import single cell data
- Choose the filtered HDF5 file for the MALT sample produced by Cell Ranger
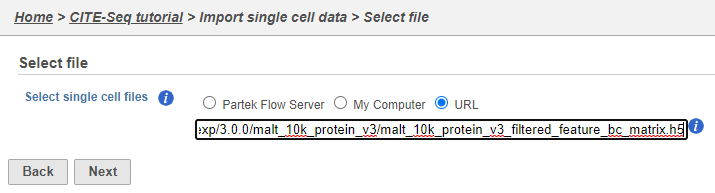
If you have already moved the .h5 file to where Partek Flow is installed, choose the Partek Flow Server radio button and browse to its location. If you have downloaded the .h5 file to your local machine, choose the My computer radio button and upload the file through your web browser. Otherwise, select the URL radio button and paste in the following link (Figure 2):
http://cf.10xgenomics.com/samples/cell-exp/3.0.0/malt_10k_protein_v3/malt_10k_protein_v3_filtered_feature_bc_matrix.h5
- Click Next
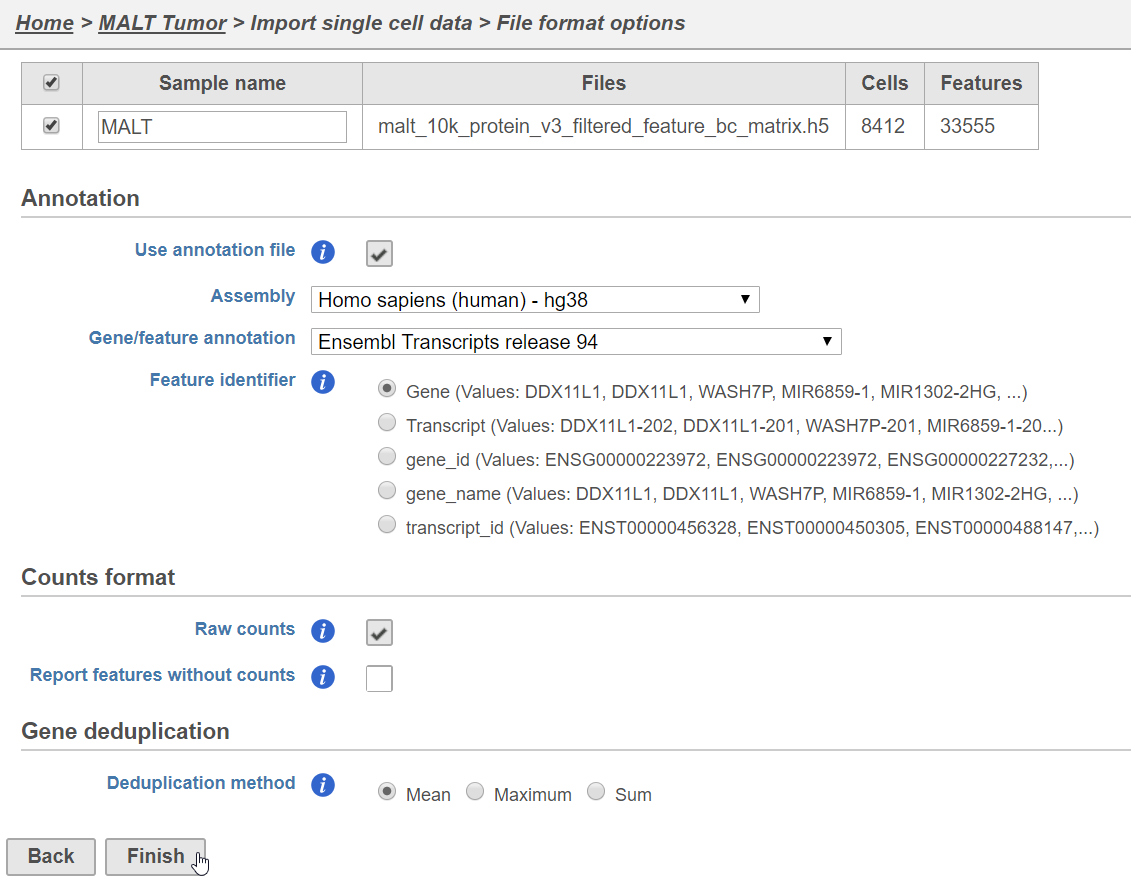
- Name the sample MALT (the default is the file name)
- Specify the annotation used for the gene expression data (here, we choose Homo sapiens (human) - hg38 and Ensembl Transcripts release 94). If Ensembl 94 is not available from the drop-down list, choose Add annotation and download it.
- Uncheck Report features without counts
- Click Finish (Figure 3)
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |