Create a new Project
Let's start by creating a new project.
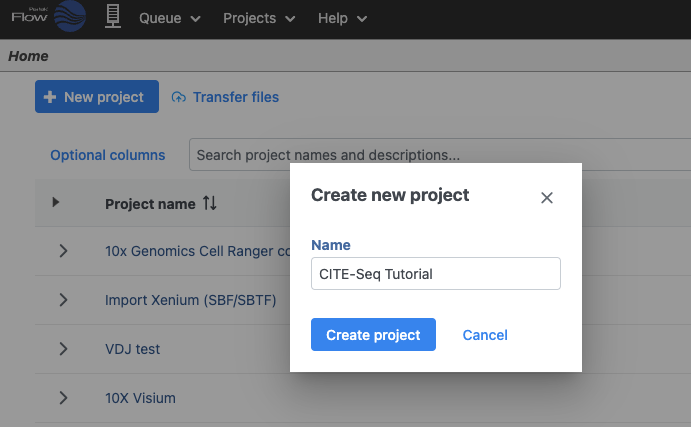
- On the Home page, click New project (Figure 1)
- Give the project a name
- Click Create project
Import data
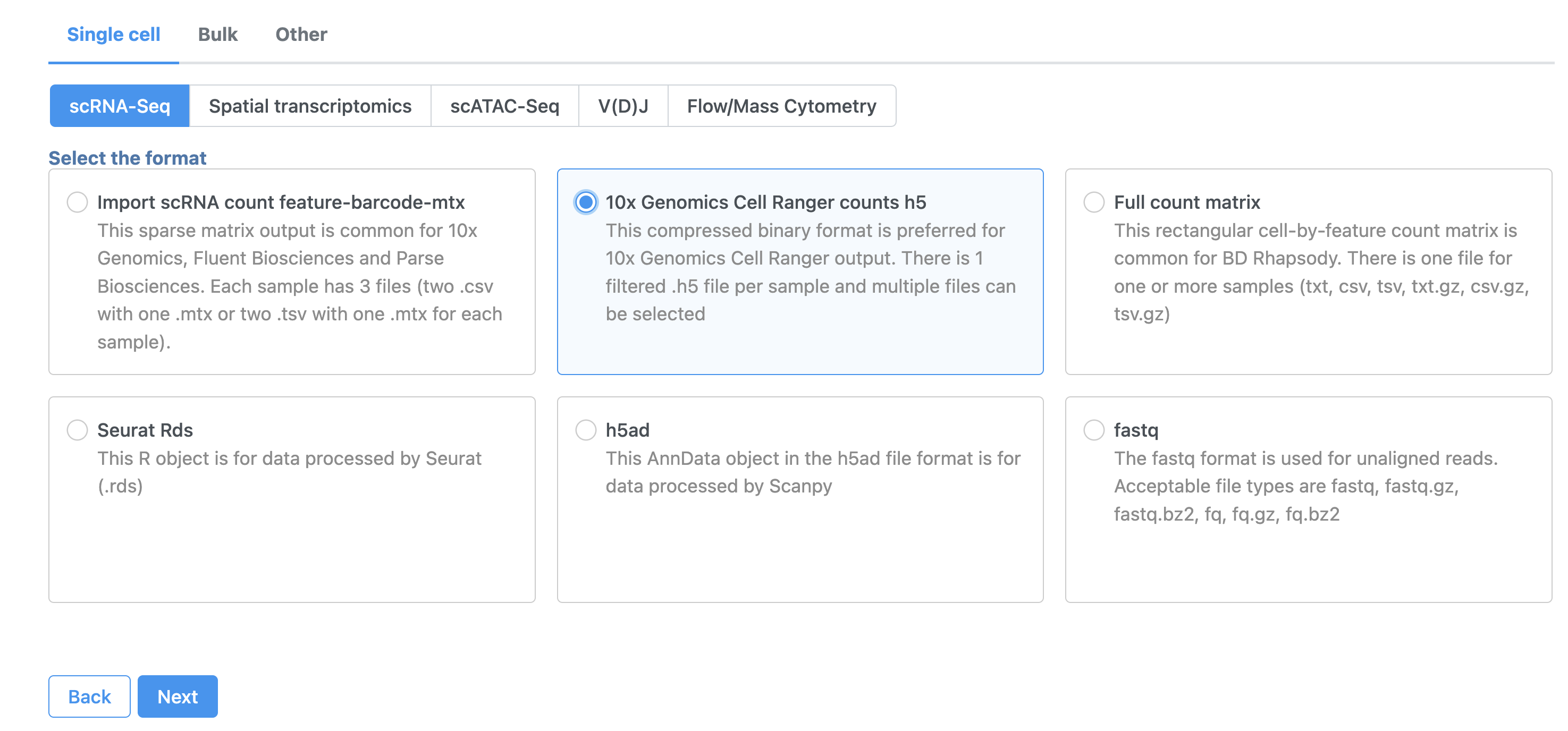
- In the Analyses tab, click Add data
- Click 10x Genomics Cell Ranger counts h5 (Figure 2)
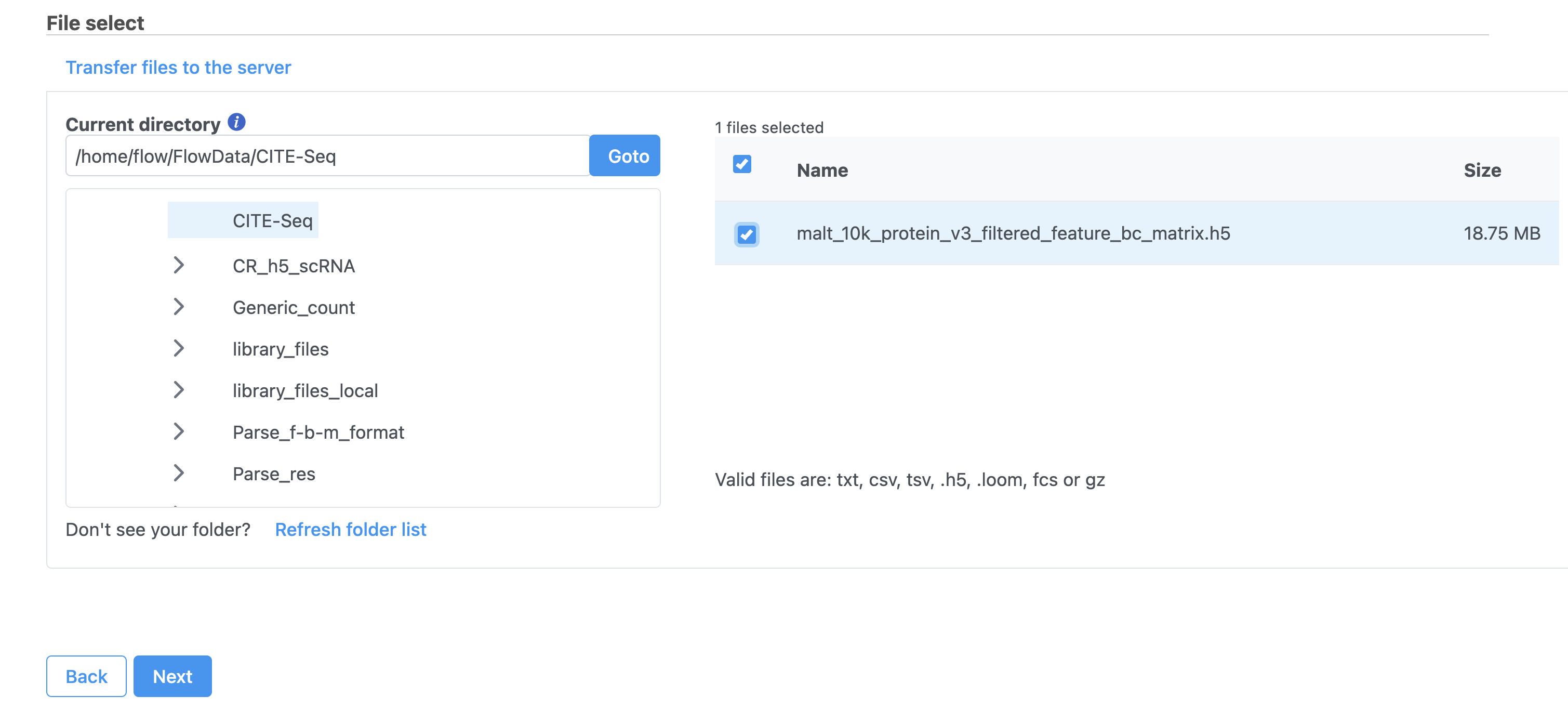
- Choose the filtered HDF5 file for the MALT sample produced by Cell Ranger
- Click Next
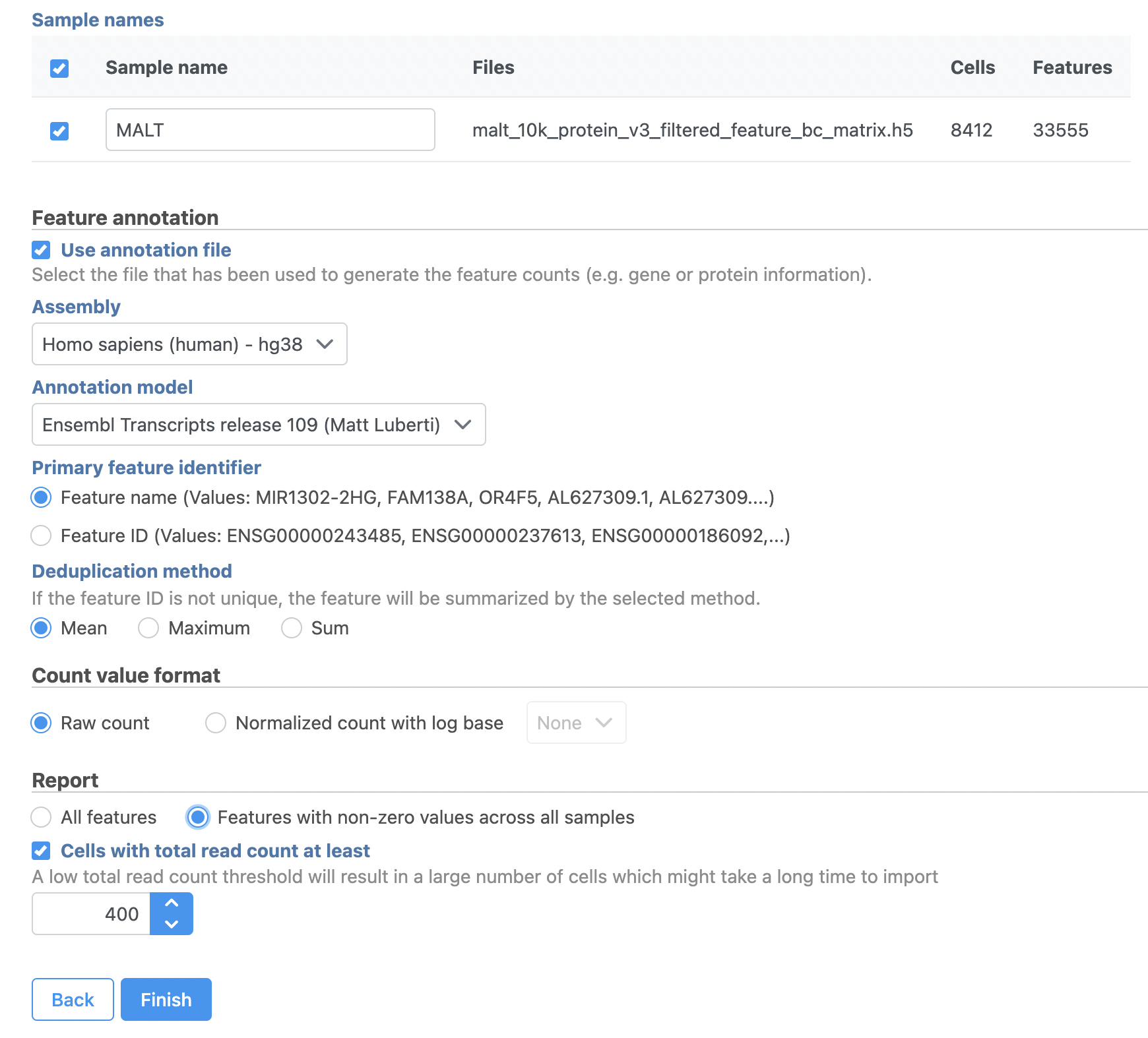
- Name the sample MALT (the default is the file name)
- Specify the annotation used for the gene expression data (here, we choose Homo sapiens (human) - hg38 and Ensembl Transcripts release 109). If Ensembl 109 is not available from the drop-down list, choose Add annotation and download it.
- Check Features with non-zero values across all samples in the Report section
- Click Finish (Figure 3)
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
7 | rates |