Next, we will perform some exploratory analysis on the merged mRNA and protein expression data and visualize the data in preparation to identify cell populations.
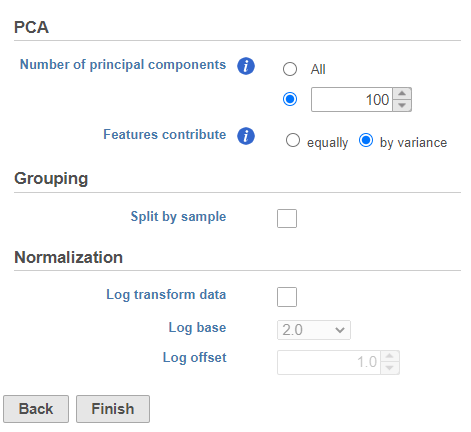
PCA
Because the merged count matrix has thousands of features, it is a good idea to reduce the dimensionality of the data for more efficient downstream processing.
- Click the Merged counts data node
- Click Exploratory analysis in the toolbox
- Click PCA
- Click Finish to run the PCA with default settings (Figure ?)
- Double click the PCA data node to open the task report
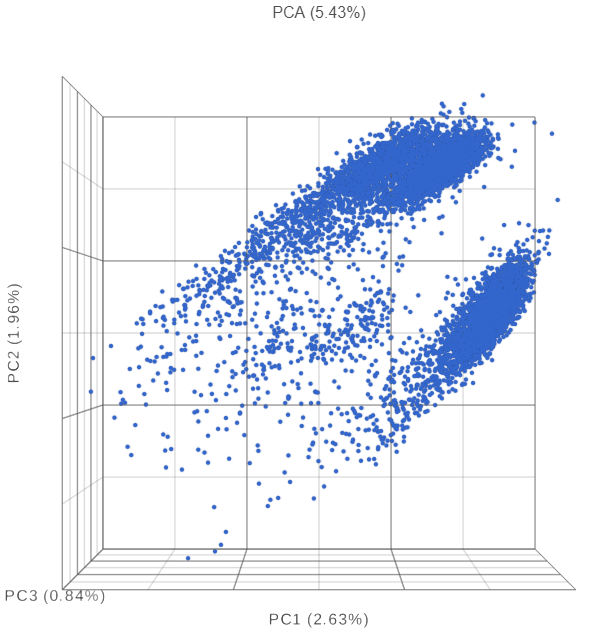
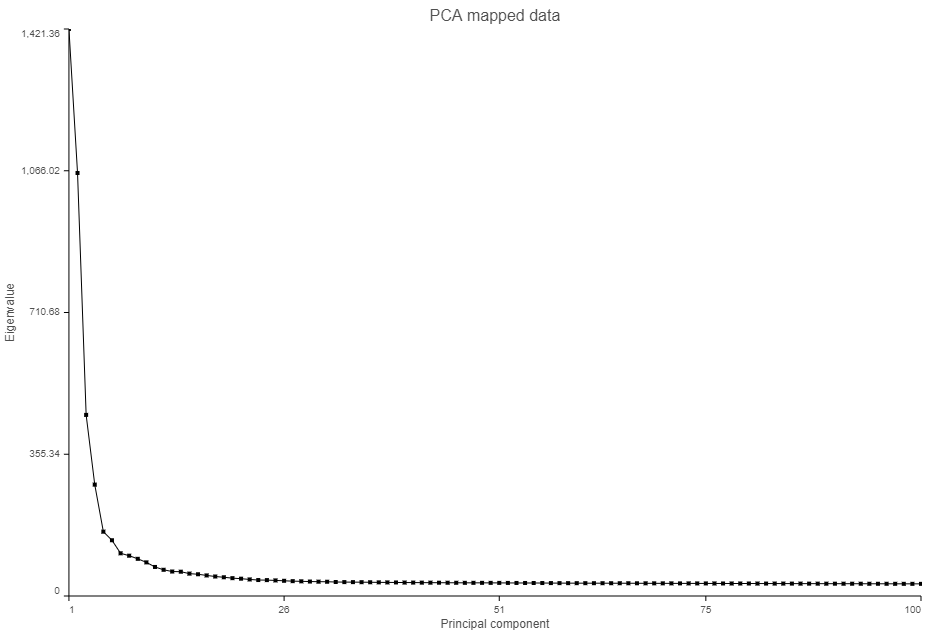
The PCA plot will open in a new data viewer session. A 3D scatterplot will be displayed on the canvas (Figure ?)
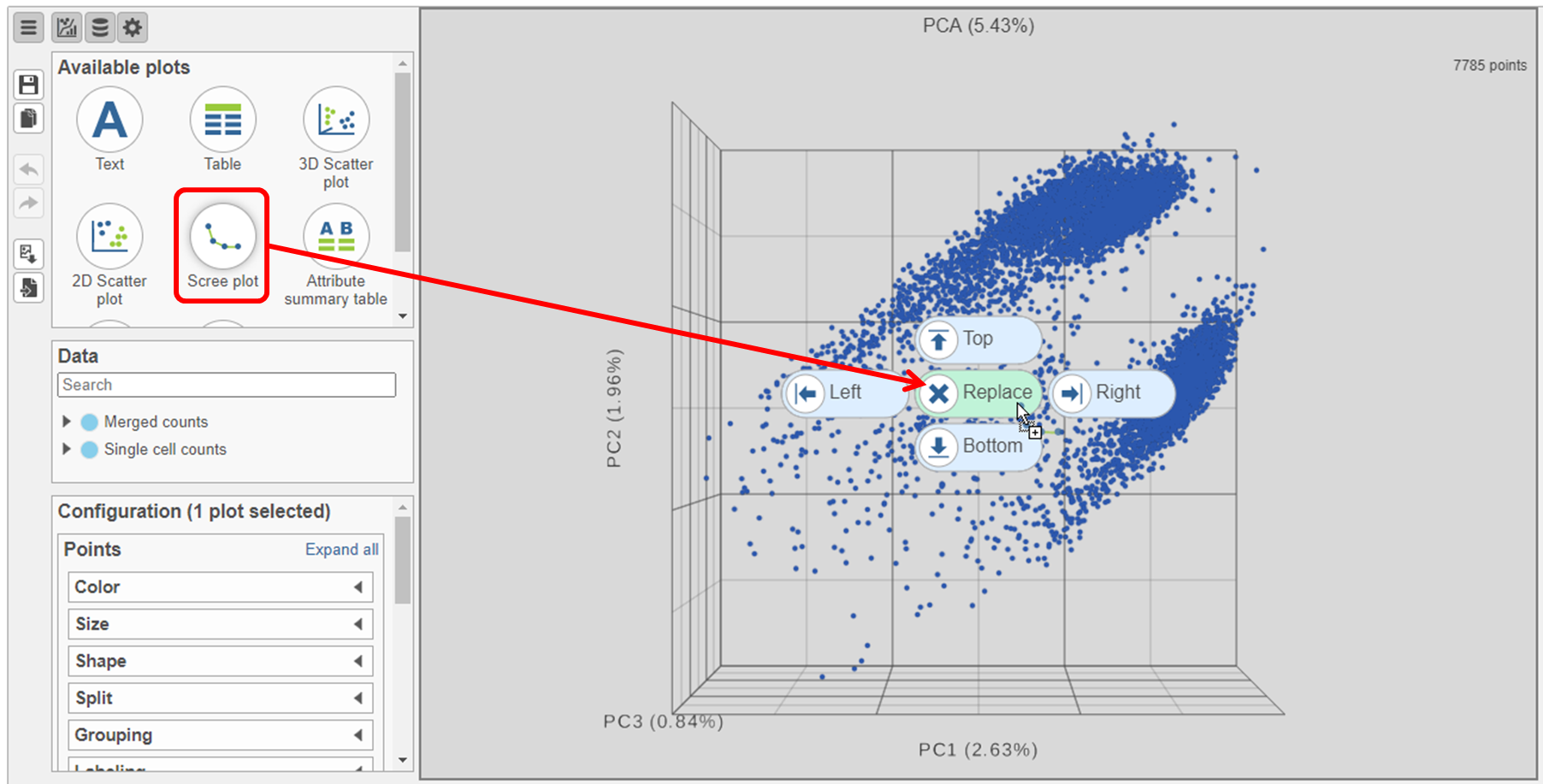
- Click and drag the Scree plot from the Available plots card on the left onto the canvas
- Drop it over the Replace option (Figure ?)
- Select PCA as data for the new Scree plot
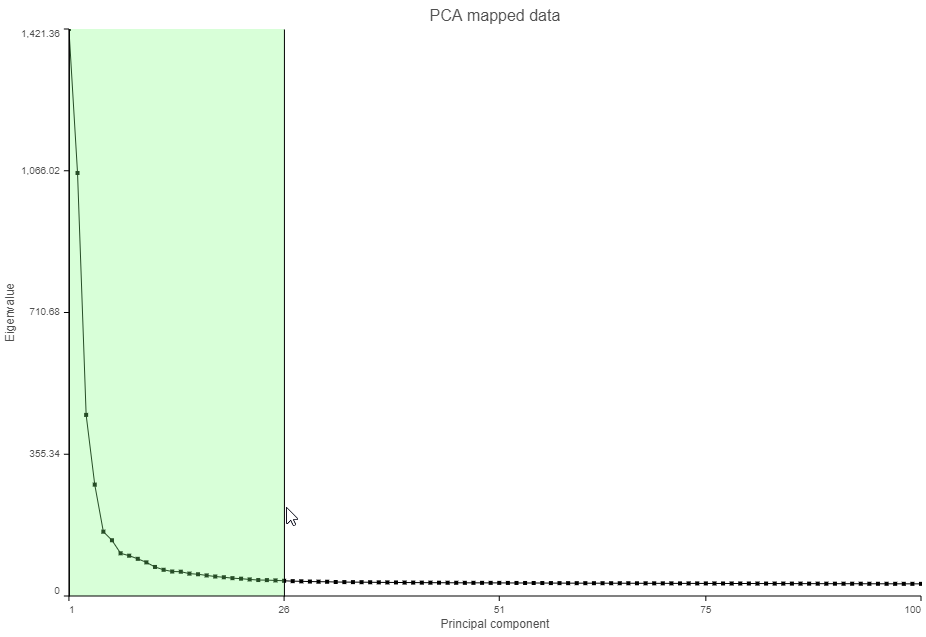
- Click and drag over the first set of PCs to zoom in (Figure ?)
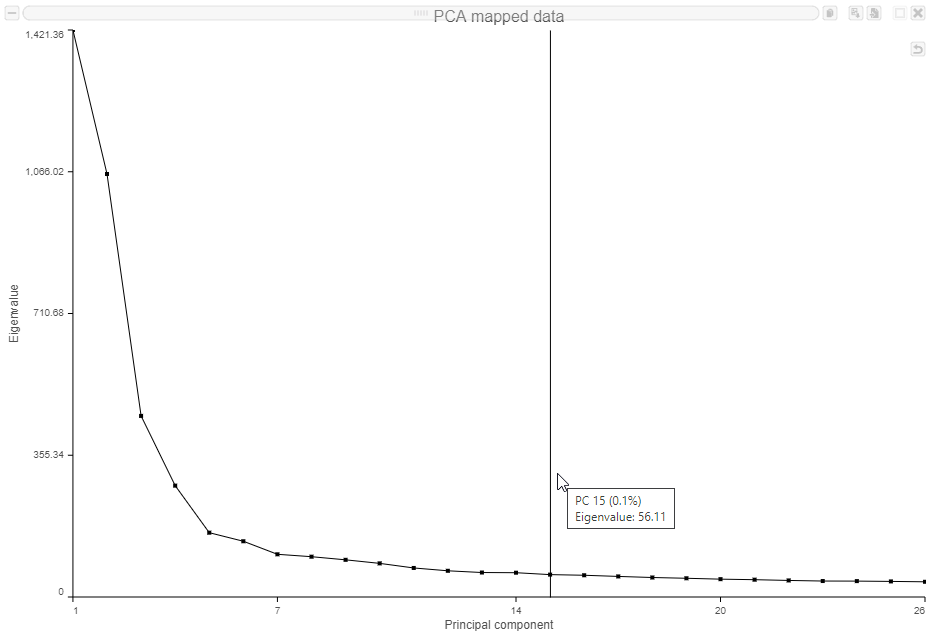
- Mouse over the Scree plot to identify the point where additional PCs offer little additional information (Figure ?)
In this data set, a reasonable cut-off could be set anywhere between around 10 and 30 PCs. We will use 15 in downstream steps.
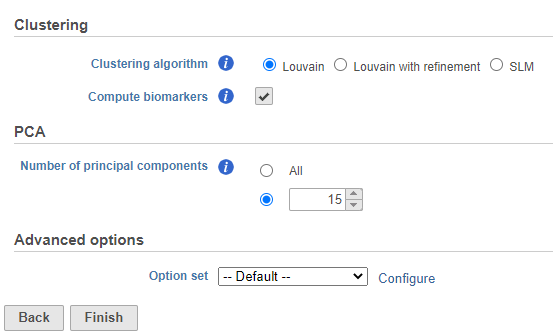
Graph-based clustering
We can use Graph-based clustering to group similar cells together in an unsupervised manner.
- Click the project name near the top to go back to the Analyses tab
- Click the circular PCA data node
- Click Exploratory analysis in the toolbox
- Click Graph-based clustering
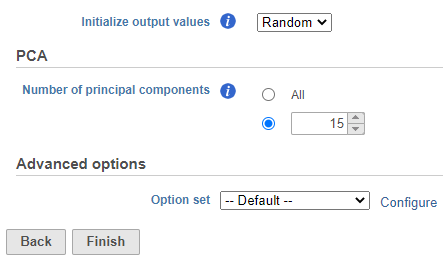
- Set the number of principal components to 15 (Figure ?)
- Click Finish to run the task
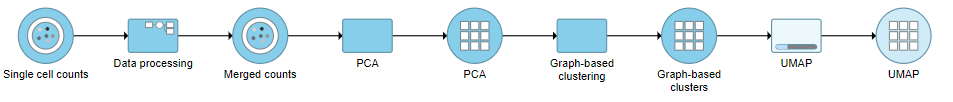
UMAP
Once the graph-based clustering task hs completed, we can visualize the results with a UMAP plot.
- Click the circular Graph-based clusters data node
- Click Exploratory analysis in the toolbox
- Click UMAP
- Set the number of principal components to 15 (Figure ?)
- Click Finish to run the task
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |