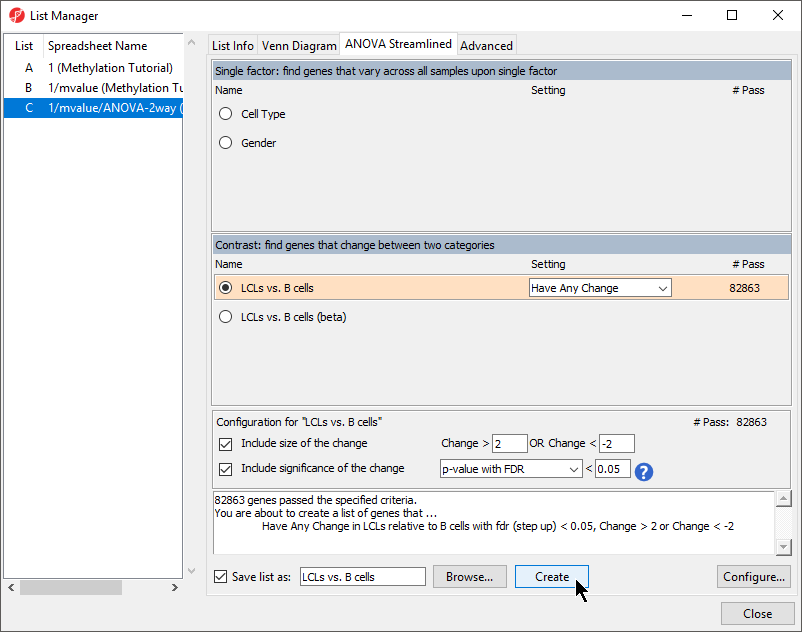
To analyze differences in methylation between our experimental groups, we need to create a list of deferentially methylated loci.
- Select Create Marker List from the Analysis section of the Illumina BeadArray Methylation workflow
- Select LCLs vs. B cells (Figure 1)
Figure 1. Creating a list of significantly differentially methylated loci
- Leave Include size of the change selected and set to Change > 2 OR Change < -2
- Leave Include significance of the change selected and set to p-value with FDR < 0.05
- Select Create
- Select Close to exit the list manager
The new spreadsheet LCLs vs. B cells (LCLs vs. B cells) will open in the Analysis tab. You may want to save the project before proceeding to the next section of the tutorial.
- Select File from the main taskbar
- Select Save Project...
- Give the project a name; we chose Methylation Tutorial
- Select Save
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
13 | rates |
Overview
Content Tools