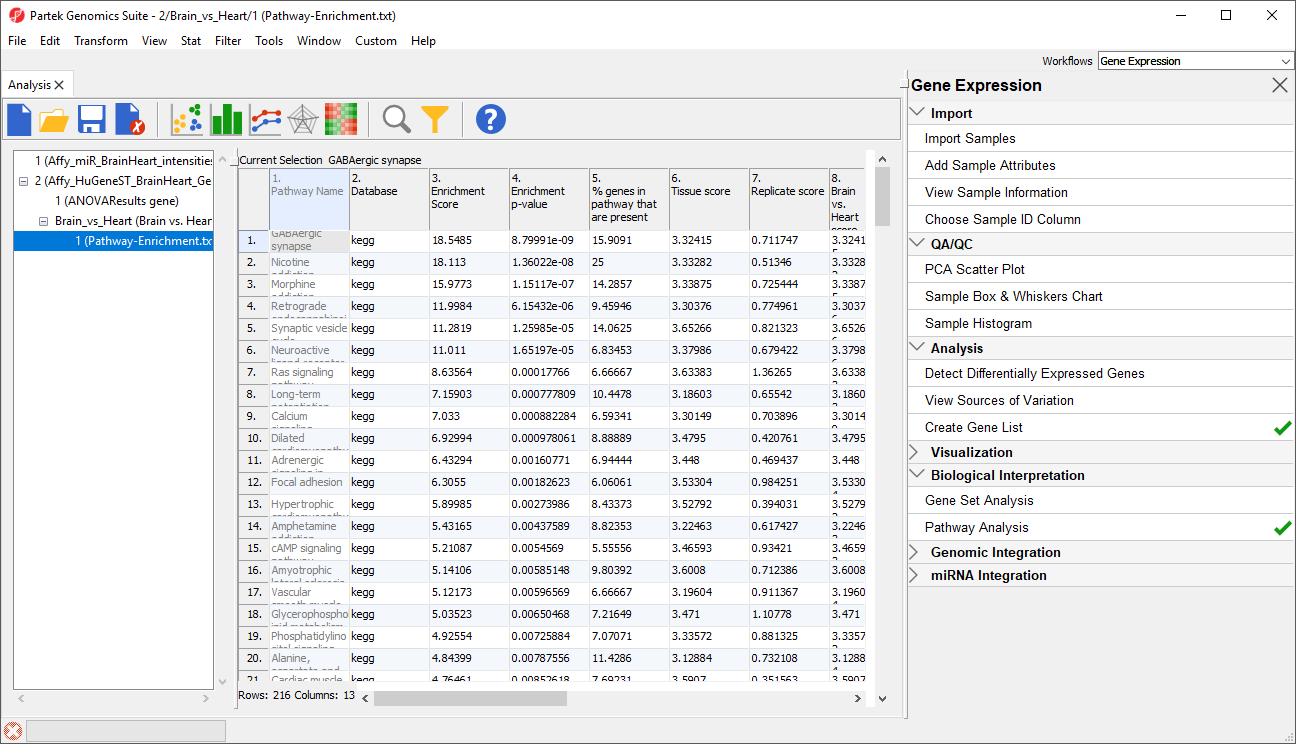
Pathway enrichment generates a results spreadsheet, Pathway-Enrichment.txt, visible in both Partek Genomics Suite (Figure 1) and in Partek Pathway.
Contents of the pathway enrichment spreadsheet
The spreadsheet includes 13 columns with information for each pathway represented in the source gene list.
1. Pathway Name - the name of the KEGG pathway
2. Database - the source database for the pathway annotation
3. Enrichment score - the negative natural log of the enrichment p-value derived from the contingency table (Fisher's Exact test) or the Chi-squared test
4. Enrichment p-value - the enrichment p-value derived from the contingency table (Fisher's Exact test) or the Chi-squared test
5. % genes in pathway that are present - the percentage of genes from the pathway that are present in the source gene list
6. Tissue score, 7. Replicate score, 8. Brain vs. Heart score - for each factor, interaction, and contrast in the ANVOA results spreadsheet, a separate score is calculated. This is derived form the negative log (base 10) of the average p-value for genes within the pathway for each factor. A high score indicates that the genes that fall into the pathway have a low p-value for the given factor.
9. # genes in list, in pathway - number of genes from the list in the pathway
10. # genes not in list, in pathway - number of genes from the pathway, not in the list
11. # genes in list, not in pathway - number of genes in list, not in the pathway
12. # genes, not in list, not in pathway - number of genes not in the pathway or the list that are included in KEGG database pathways for the species
13. Pathway ID - KEGG pathway ID
Tasks available in Partek Genomics Suite
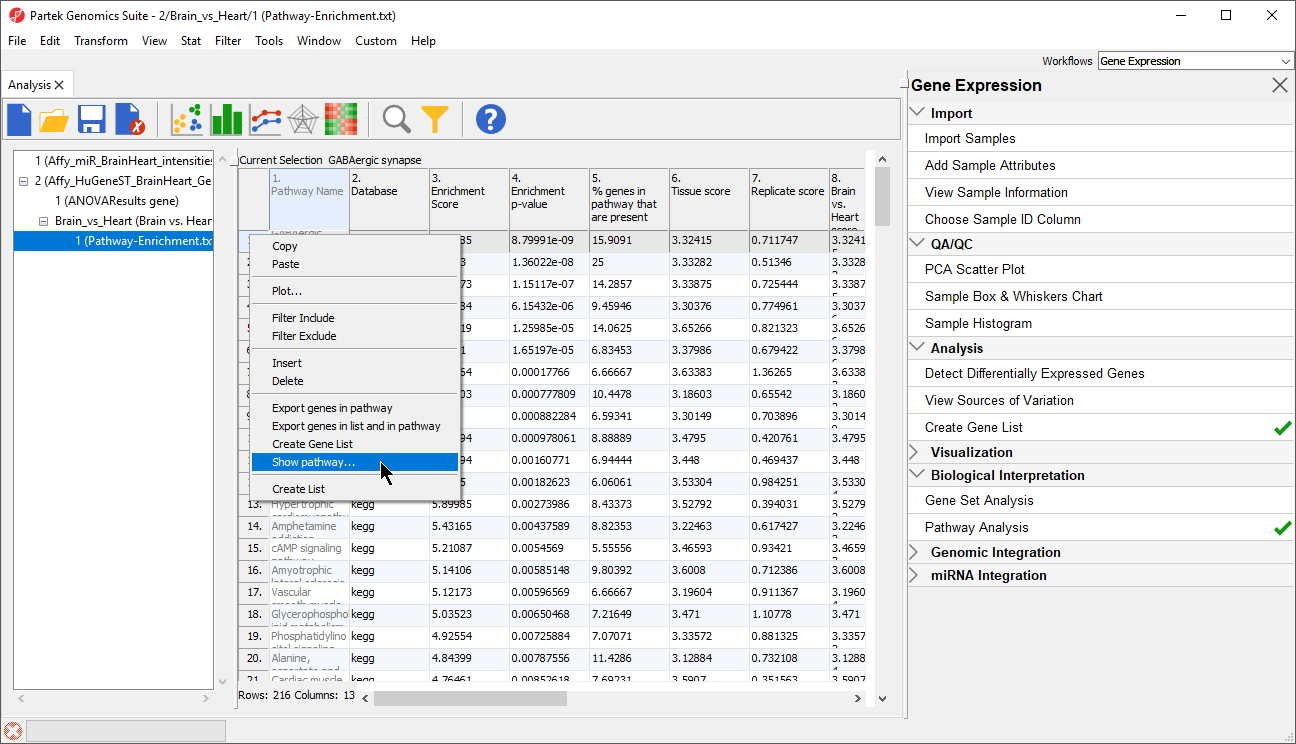
In Partek Genomics Suite, we can view several new options that are available for each pathway (row) in the Pathway-Enrichment.txt spreadsheet.
- Right-click the row header of any row in the Pathway-Enrichment.txt spreadsheet (Figure 2)
The new options include:
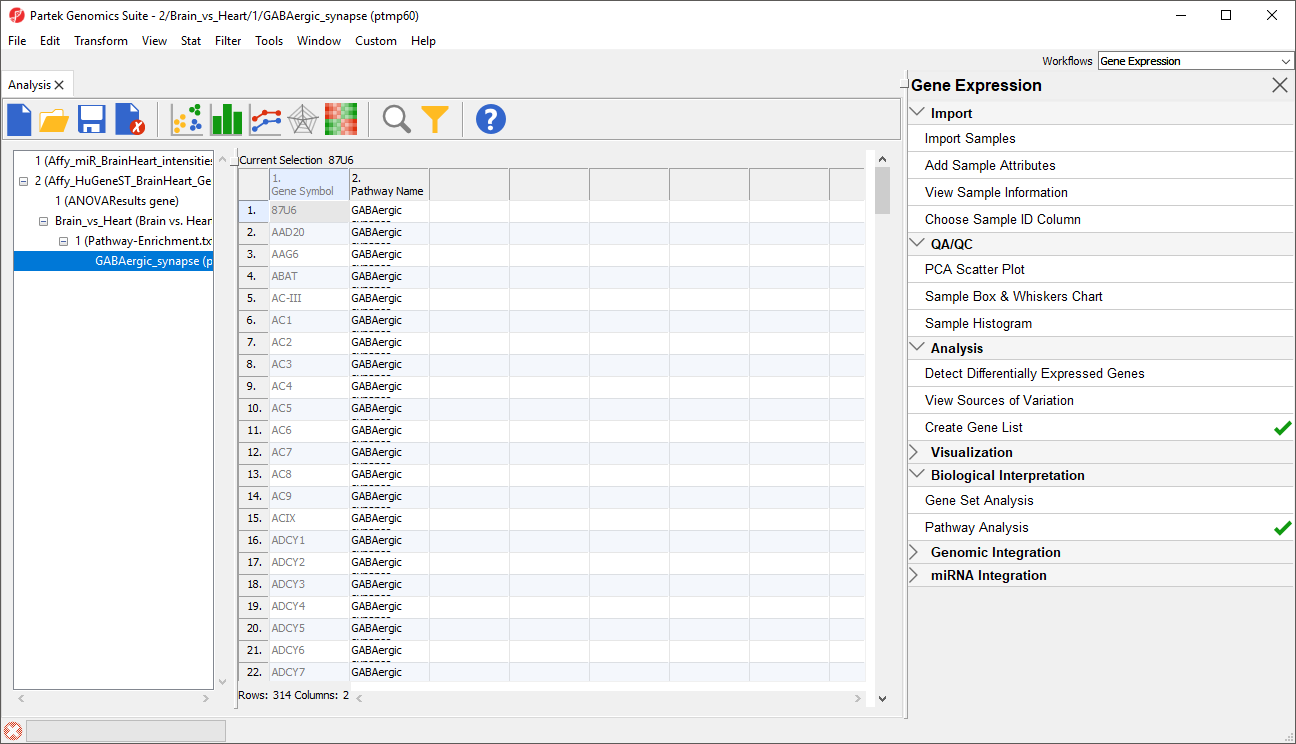
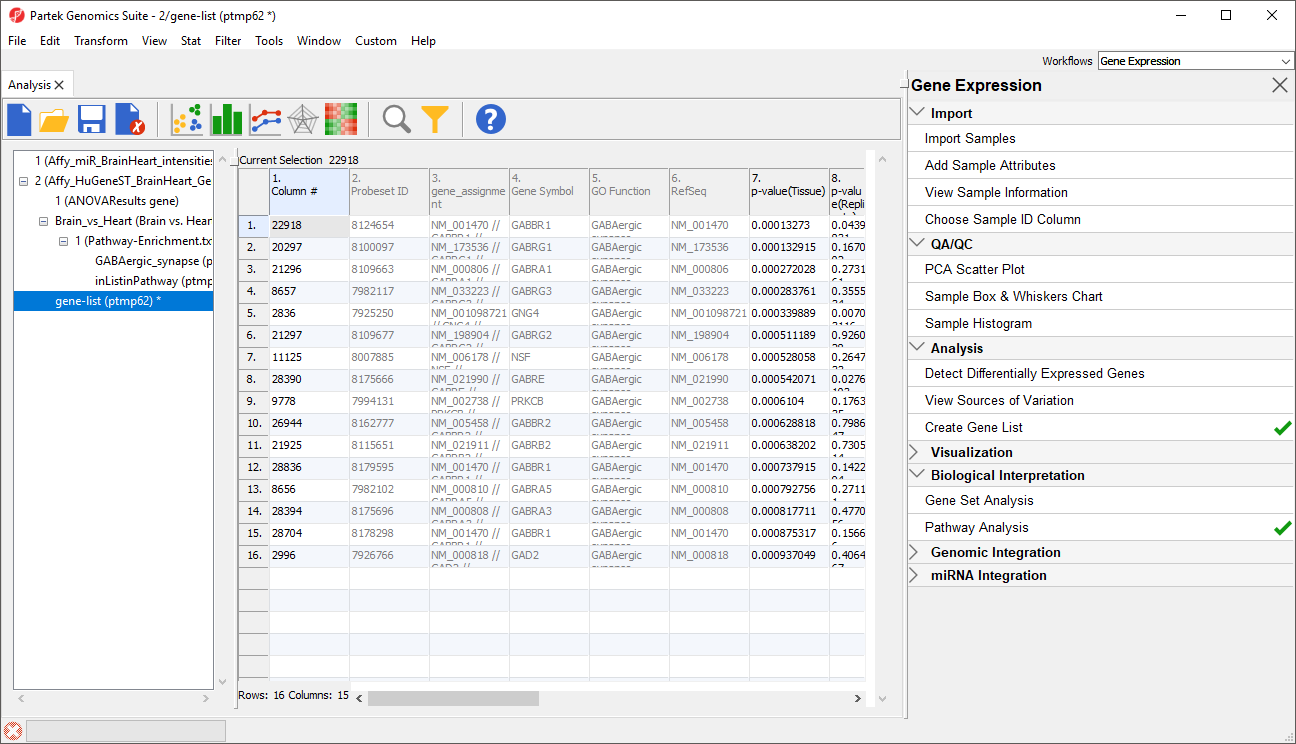
Export genes in pathway, which creates a child spreadsheet ofPathway-Enrichment.txt that contains all the genes from the selected pathway(s) (Figure 3). This new spreadsheet includes gene symbols and their pathway.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
10 | rates |