To detect differential methylation between CpG loci in different experimental groups, we can perform an ANOVA test. For this tutorial, we will perform a simple one-way ANOVA to compare the methylation states of the four experimental groups.
- Select Defect Differential Methylation from the Analysis section of the Illumina BeadArray Methylation workflow
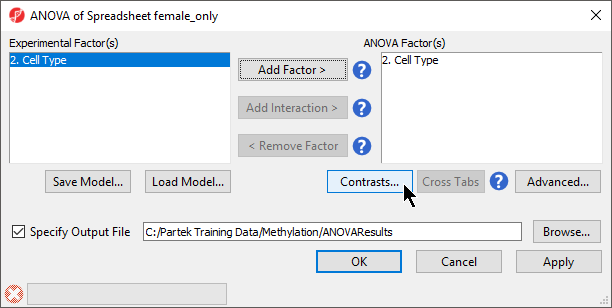
- Select 2. Cell Type from the Experimental Factor(s) panel
- Select Add Factor > to move 2. Cell Type to the ANOVA Factor(s) panel (Figure 1)
- Select Contrasts...
- Select Yes for Data is already log transformed? because M-values are based on logit transformation
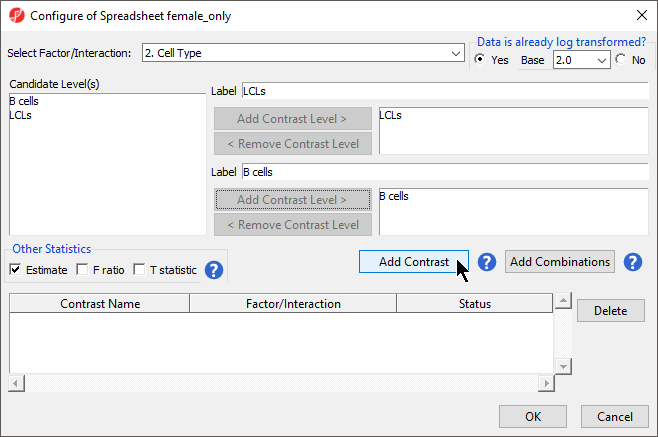
- Select LCLs
- Select Add Contrast Level > for the upper group
- Select B cells
- Select Add Contrast Level > for the lower group
- Select the Estimate box in the Other Statistics section of the Configure dialog (Figure 2)
- Select Add Contrast
By default, the fold-change value for each contrast will be calculated with the upper group as the numerator and the lower group as the denominator. Selecting Estimate will also include the difference in methylation levels between the groups at each CpG site in the output. These values will be needed later in the tutorial.
- Select OK to close the Configuration dialog
The Contrasts... button of the ANOVA dialog now reads Contrasts Included
- Select OK to close the ANOVA dialog and run the ANOVA
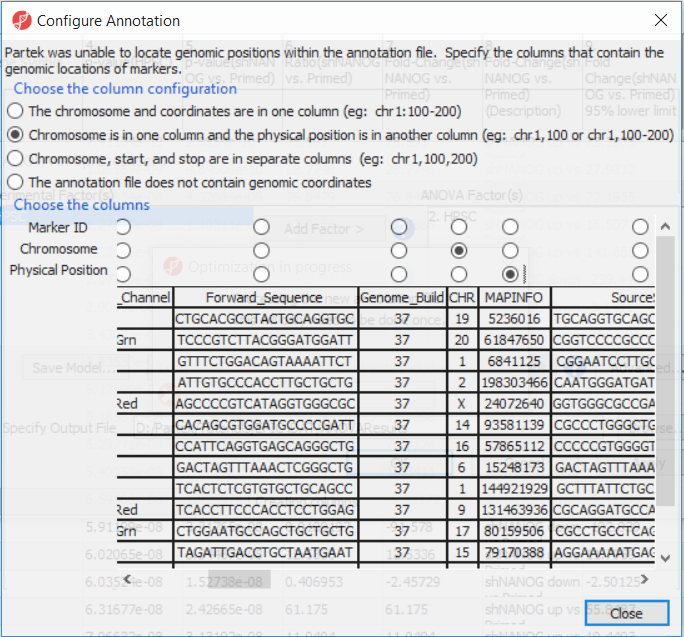
If this is the first time you have analyzed a MethylationEPIC array using the Partek Genomics Suite software, the manifest file may need to be configured. If it needs configuration, the Configure Annotation dialog will appear (Figure 3).
- Select Chromosome is in one column and the physical location is in another column for Choose the column configuration
- Select Ilmn ID for Marker ID
- Select CHR for Chromosome i
- Select MAPINFO for Physical Position
- Select Close
This enables Partek Genomics Suite to parse out probe annotations from the manifest file.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
1 | rates |