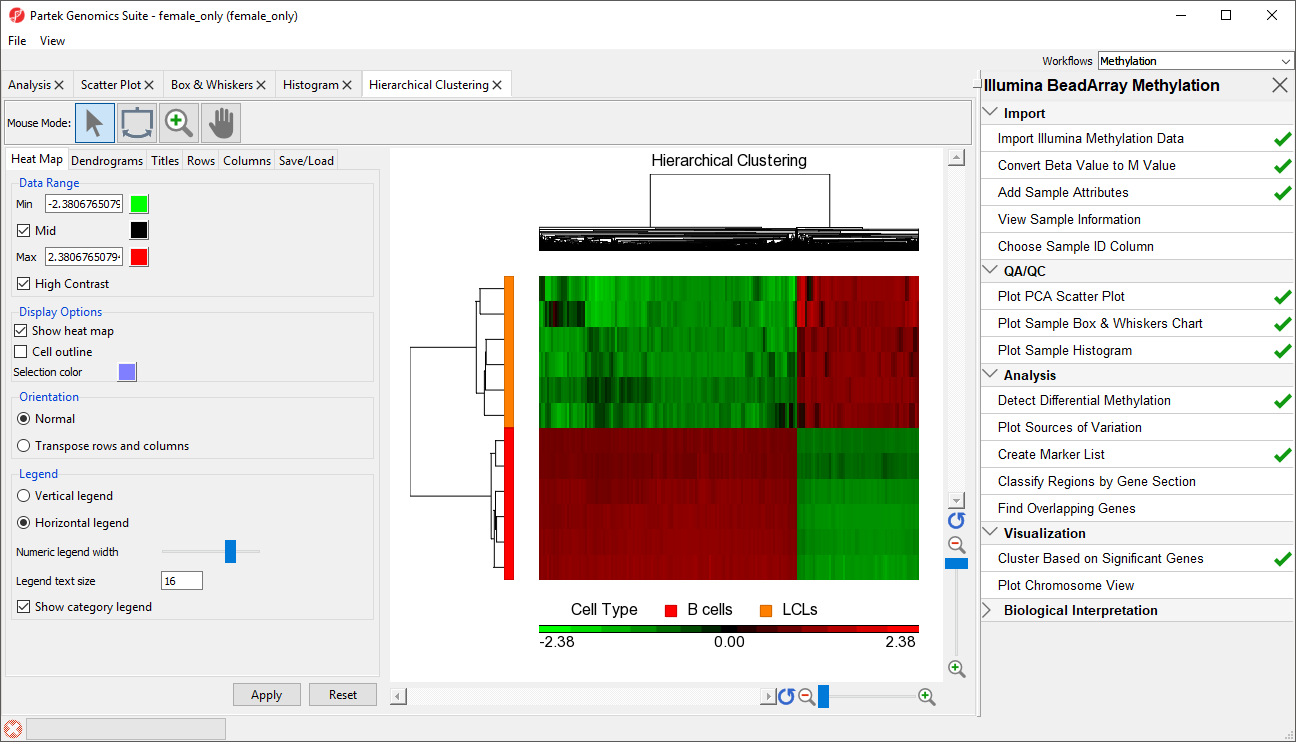
The significant CpG loci detected in the previous step actually form a methylation signature that differentiates , which we shall now visualise by a heat map.
- Select the (LCLs vs. B cells) spreadsheet in the spreadsheet pane on the left
- Select Cluster Based on Significant Genes from the Visualization panel of the Illumina BeadArray Methylation workflow
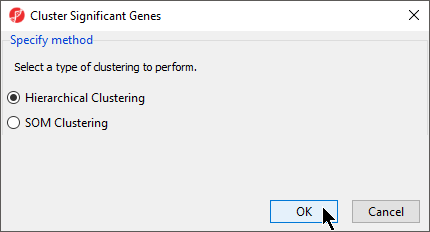
- Select Hierarchical Clustering for Specify Method (Figure 1)
- Select OK
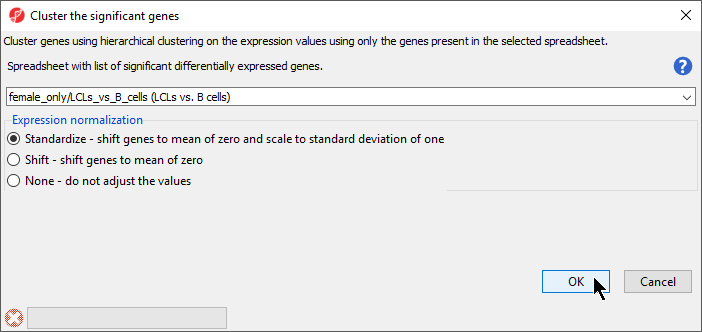
- Verify that (CpG of interest) is selected in the drop-down menu
- Select Standardize for Expression normalization (Figure 2)
- Select OK
The heat map will be displayed on the Hierarchical Clustering tab (Figure 3).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |