To analyze differences in methylation between our experimental groups, we need to create a list of deferentially methylated loci.
- Select Create Marker List from the Analysis section of the Illumina BeadArray Methylation workflow
- Select 1/anova-1way_autosomal_only (ANOVA-1way_autosomal_only) as the source spreadsheet from the left-hand panel
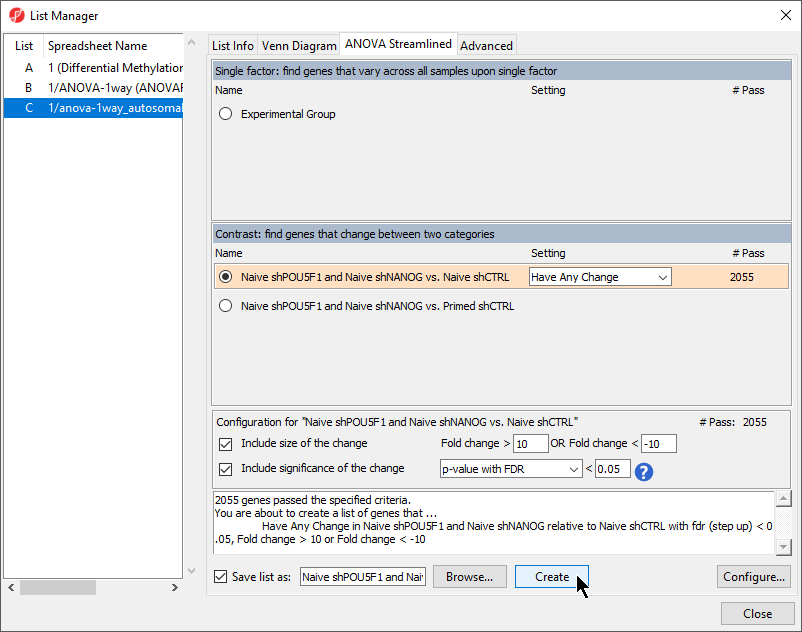
- Select Naive shPOU5F1 and Naive shNANOG vs. Naive shCTRL from the Contrast: find genes that change between two categories section of the ANOVA Streamlined tab of the List Manager dialog
- Set Fold change > to 10
- Set OR Fold change < to -10
- Leave the rest of the option set to defaults (Figure 1)
Figure 1. Creating a marker list of differentially methylated genes between Naive shPOU5F1 and Naive shNANOG vs. Naive shCTRL
- Select Create
- Create another marker list with the same criteria for the contrast Naive shPOU5F1 and Naive shNANOG vs. Primed shCTRL
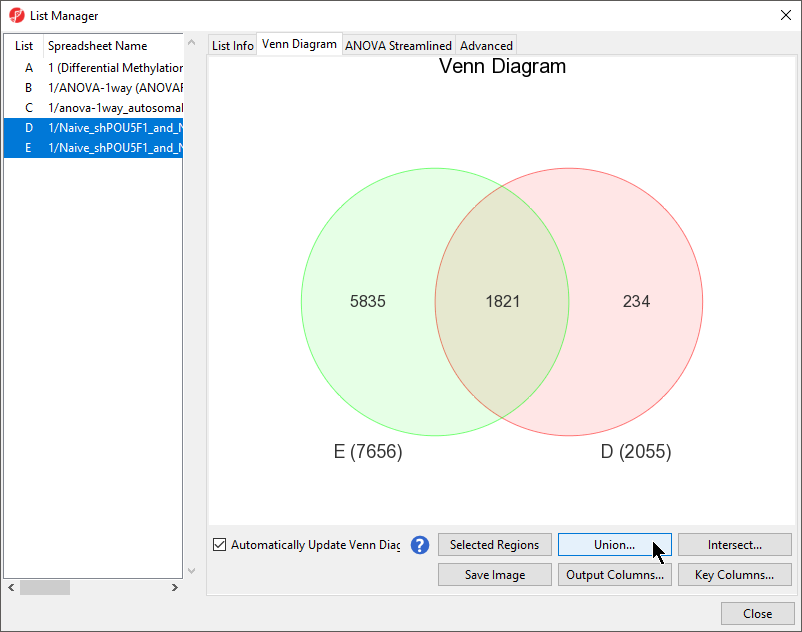
- Select the Ven Diagram tab on the List Manager dialog
- Select both D and E from as the source spreadsheets from the left-hand panel list by holding Ctrl and selecting both D and E
- Select Union... to create a list with probes from both spreadsheets (Figure 2)
Figure 2. Creating a marker list with markers from two spreadsheets
- Select Close to exit the List Manager dialog
- Select () from the quick action bar to save the newly created spreadsheet
- Specify a name for the spreadsheet, we chose CpG Loci of Interest, using the Save File dialog
- Select Save to save the spreadsheet
You may want to save the project before proceeding to the next section of the tutorial.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |
Overview
Content Tools