To detect differential methylation between CpG loci in different experimental groups, we can perform an ANOVA test. For this tutorial, we will perform a simple one-way ANOVA to compare the methylation states of the four experimental groups.
- Select Defect Differential Methylation from the Analysis section of the Illumina BeadArray Methylation workflow
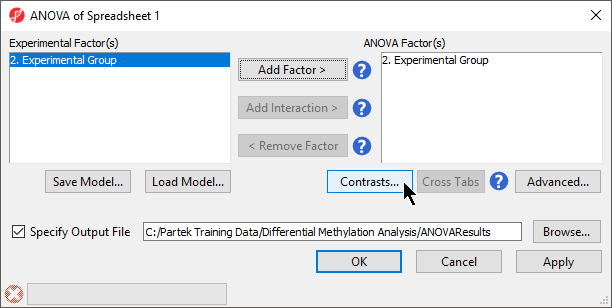
- Select 3. shRNA treatment from the Experimental Factor(s) panel
- Select Add Factor > to move 3. shRNA treatment to the ANOVA Factor(s) panel (Figure 1)
- Select Contrasts...
- Select Yes for Data is already log transformed? because M-values are based on logit transformation
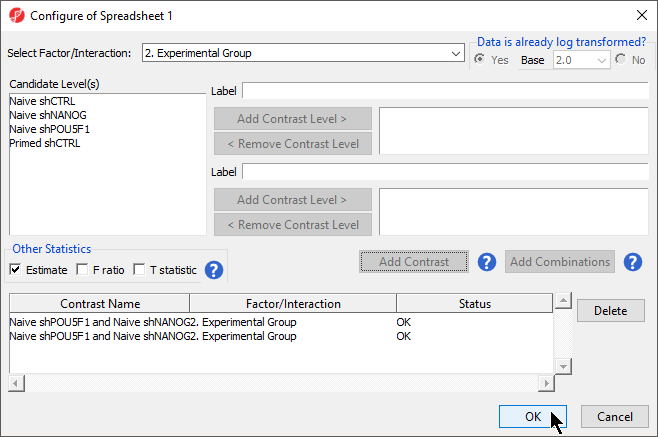
- Select Naive shPOU5F1
- Select Add Contrast Level > for the upper group
- Repeat to add Naive shNANOG to the upper group
- Select Naive shCTRL
- Select Add Contrast Level > for the lower group
- Select Add Contrast
- Repeat steps to create an additional contrast: Naive shPOU5F1 and Naive shNANOG vs. Primed shCTRL
- Select the Estimate box in the Other Statistics section of the Configure dialog (Figure 2)
By default, the fold-change value for each contrast will be calculated. Selecting Estimate will also include the difference in methylation levels between the groups at each CpG site in the output. These values will be needed later in the tutorial when we filter the differentially methylated loci.
- Select Add Combination
- Select OK to close the Configuration dialog
The Contrasts... button of the ANOVA dialog now reads Contrasts Included
- Select OK to close the ANOVA dialog and run the ANOVA
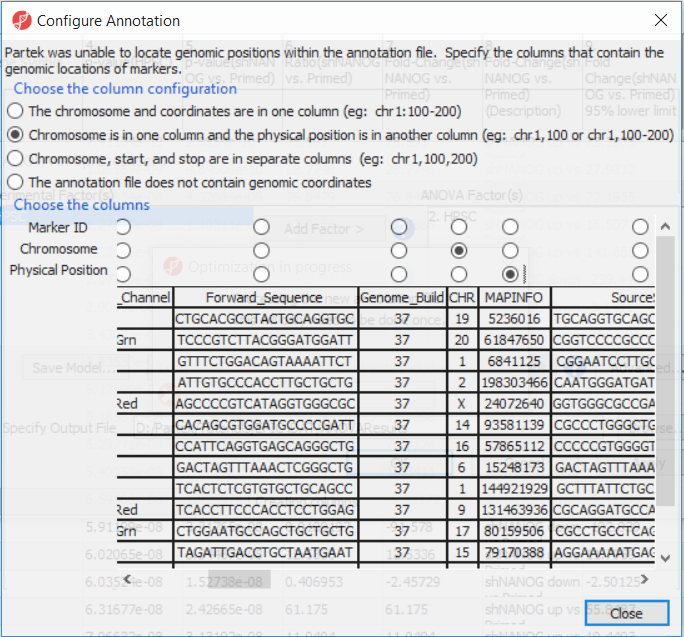
If this is the first time you have analyzed a MethylationEPIC array using the Partek Genomics Suite software, the manifest file may need to be configured. If it needs configuration, the Configure Annotation dialog will appear (Figure 3).
- Select Chromosome is in one column and the physical location is in another column for Choose the column configuration
- Select Ilmn ID for Marker ID
- Select CHR for Chromosome i
- Select MAPINFO for Physical Position
- Select Close
Select Close. This enable Partek Genomics Suite to parse out probe annotation from the manifest file.
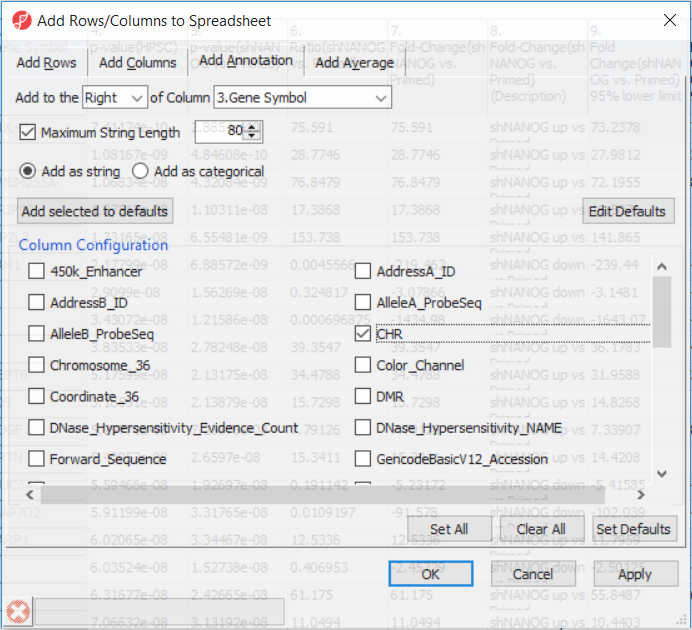
Going forward, analysis of differentially methylated loci typically includes removal of the probes on X and Y chromosomes (to avoid the problems with inactivation of one X chromosome). To annotate the ANOVA spreadsheet with the information required for filtering, right-click on the Gene Symbol column, select Insert Annotation, tick-mark the CHR filed (Figure 6) and push OK. A new column will be appended to the spreadsheet.
To enable filtering right-click on the header of the CHR column > Properties and set the type to categorical (and OK). Now, activate the Interactive Filter tool () . If needed use the drop-down list to point to the CHR column. The column chart represents the number of appearances of each chromosome in the spreadsheet (i.e. the number of probes per chromosome). To remove the probes on the X and the Y chromosome left click on the two right-most columns (the pop up balloon will show you the chromosome label) and the columns will be grayed out (Figure 7).
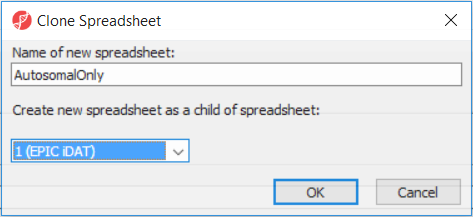
After removal of the probes on the sex chromosomes, let us extract all the autosomal probes to a new spreadsheet. Right click on the ANOVA 1-way spreadsheet > Clone... Set the Name of new spreadsheet to AutosomalOnly and make it a child of the top-level spreadsheet (EPIC iDAT) (Figure 8). Push OK. The newly created autosomalonly spreadsheet will be a new starting point for all the downstream steps.
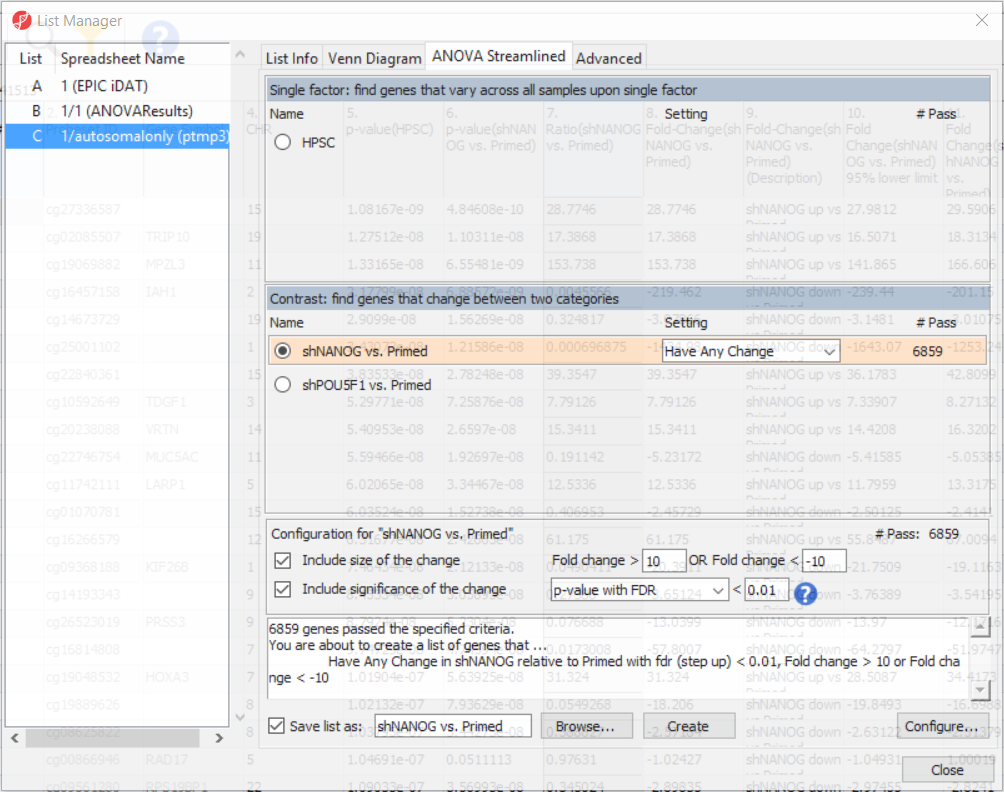
To come up with a list of differentially methylated loci, proceed to the workflow and select Create Marker List. That will open the List Manager functionality and you will be on the ANOVA Streamlined tab. Factors in the model are listed in the top section, contrasts specified in the model are in the middle, while the filter settings are at the bottom.
Select shNANOG vs Primed contrast to see the current filter configuration. By default, both fold-change and p-value with false discovery rate (FDR) correction are applied, with the number of CpG loci passing the filter given as # Pass. For this tutorial, set the fold-change to > 10 and < -10, and reduce the p-value with FDR down to 0.01 (Figure 9). Then push the Create button to save the list of significant loci under the default name (shNANOG vs. Primed). Repeat the procedure for the shPOU5F1 vs Primed, using the same cut offs.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
1 | rates |