We will be using the RNA-seq workflow to analyze RNA-seq data throughout this tutorial. The commands included in the RNA-seq workflow are also available form the command toolbar, but may be labeled differently.
- Select the RNA-seq workflow by selecting it from the Workflow drop-down menu in the upper right-hand corner of the Partek Genomics Suite window (Figure 1)
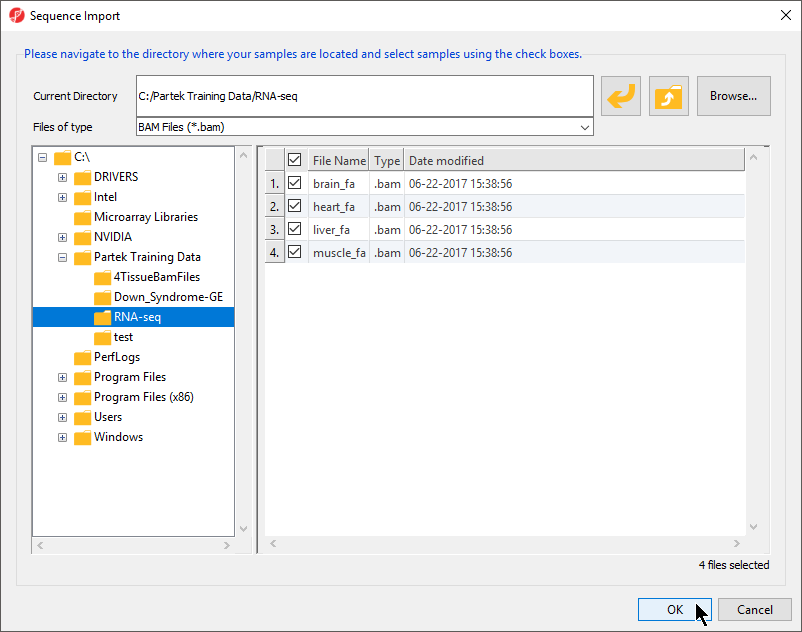
- To import the .BAM files, select Import and Manage Samples from the Import section of the RNA-seq workflow. The Sequence Import dialog box will open (Figure 2)
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |