Page History
...
- Task name - a unique name for the task
- Description (optional) - a more thorough description of the task. This text will appear when the cursor hovers over the task in the Task Menu
- Version - a unique version number for the given task name. The combination of Task name + Version must be unique (i.e., no other task already installed can have the same combination)
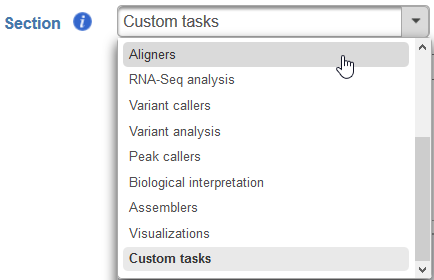
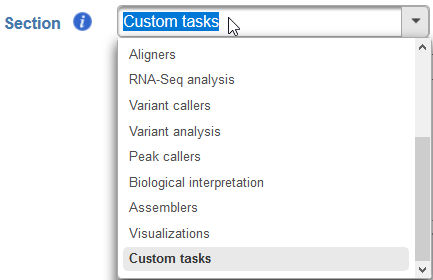
Section - choose a section in the context-sensitive Task Menu in which this task will appear or define a new section by editing the text field (Figure ?4)
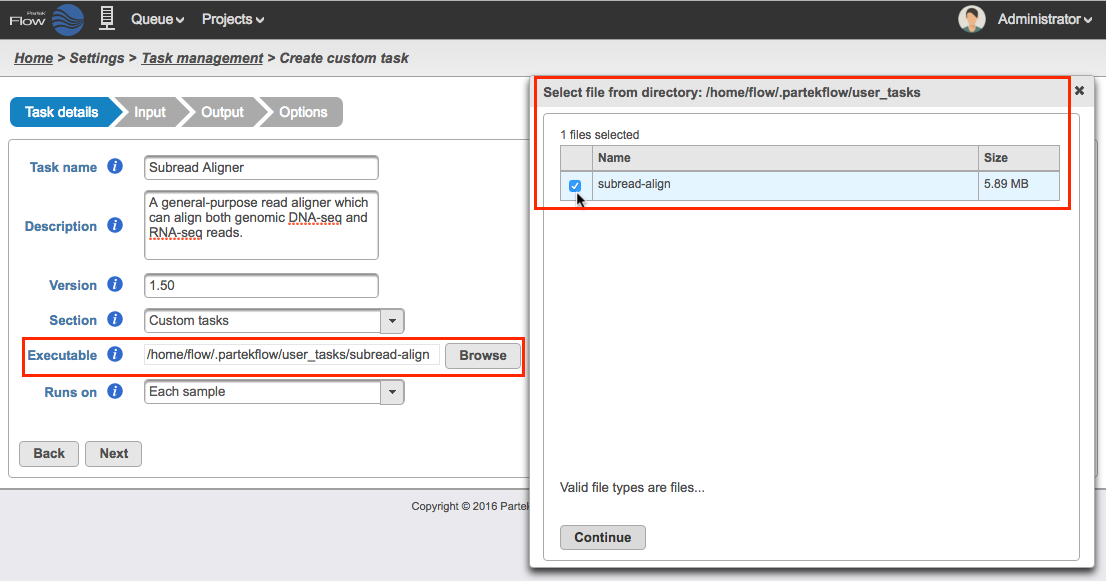
Numbered figure captions SubtitleText Choose which section in the Task Menu you want the added task to appear in. To keep all custom tasks in one section, choose 'Custom tasks' at the bottom of the drop-down list (left). To create a new section, highlight the text and manually type a new section name (right) AnchorName Choose which section in the Task Menu you want the added task to appear in. To keep all custom tasks in one section, choose 'Custom tasks' at the bottom of the drop-down list (left). To create a new section, highlight the text and manually type a new section name (right) - Executable - provide a full path to the executable that this task will run (Figure 4Figure 5) by clicking Browse. The executable is the main binary or script to call when the user added task is run. In the example in Figure 3, the full path is /home/flow/.partekflow/user_tasks. The dialog (Figure 4 5 inset) lists all the available executables stored in the directory. Select the file and click Continue to proceed.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
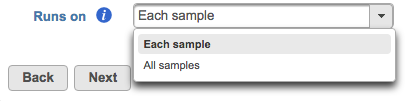
- Runs on - choose Each sample to run the executable once per individual sample (i.e., run the samples in parallel) or All samples to run the binary once with all the samples provided in a single command line (e.g, merge all samples into a single run) (Figure ?6)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
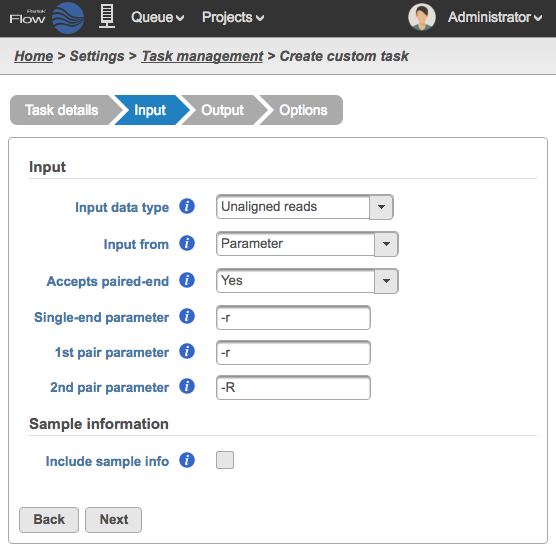
In the Input step, specify what type of data the task can run on and how to pass that data on the command line (Figure 67).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
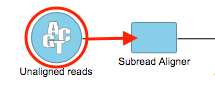
- Input data type - This is the type of data the task can be performed on. In the Analyses tab, this is the type of data node that will be selected for to the task to appear in the context-sensitive Task menu (Figure?). Partek Flow requires User added tasks to be performed on either: Unaligned reads, Aligned reads, and Variants. Additional inputs can be specified in the Options step. The type of Input data selected from the drop-down menu will determine the additional fields that need to be defined. In Figure? the Subread Aligner task accepts Unaligned reads.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Input from - choose Parameter if the input file(s) is specified using an option (e.g. -r) on the command line or Last option if the executable assumes the input file(s) is placed at the end of the command
- Accepts paired-end - choose between Yes, No or Exclusively with respect to the task's behavior with paired-end reads
- Single-end parameter - enter the parameter to add before single-end reads
- 1st pair parameter - enter the parameter to add before the first file (_1) in a paired-end reads
...
- set
- 2nd pair parameter - enter the parameter to add before the first file (_2) in a paired-end reads set
You can optionally include Sample information as an argument to pass to the command (Figure 8). Select Include sample info and enter the Sample info parameter. The parameter is used to specify the sample information file on the command line.
...
Overview
Content Tools