...
To create a new project, from the Home page click the New Project button; enter a project name and then click Create project (Figure 1). Once a new project has been created, the user is automatically directed to the Data tab of the Project Viewclick the Add data button in the Analyses tab.
| Numbered figure captions |
|---|
| SubtitleText | Transfer file in Partek Flow. |
|---|
| AnchorName | File transfer |
|---|
|
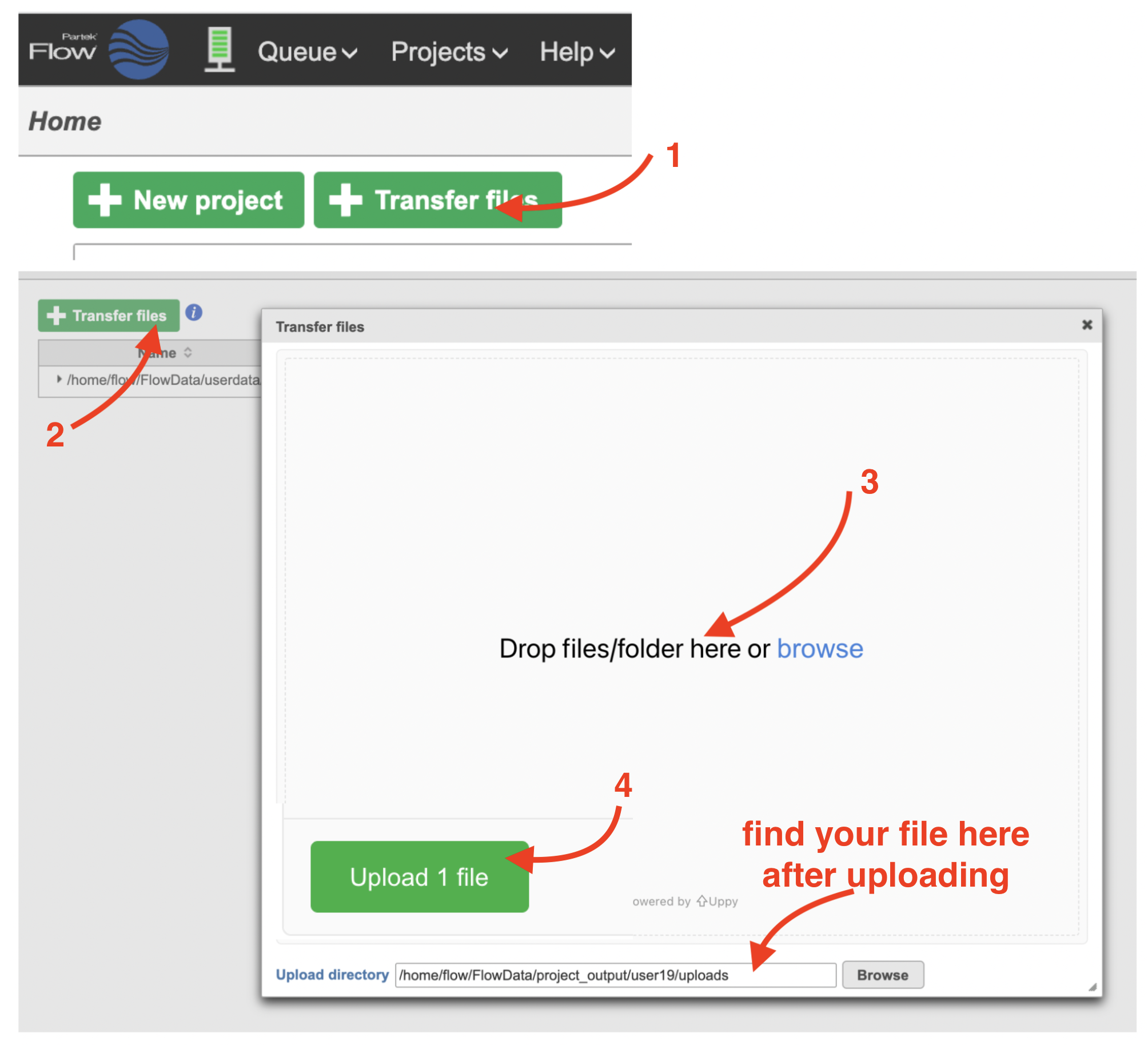 Image Removed Image Removed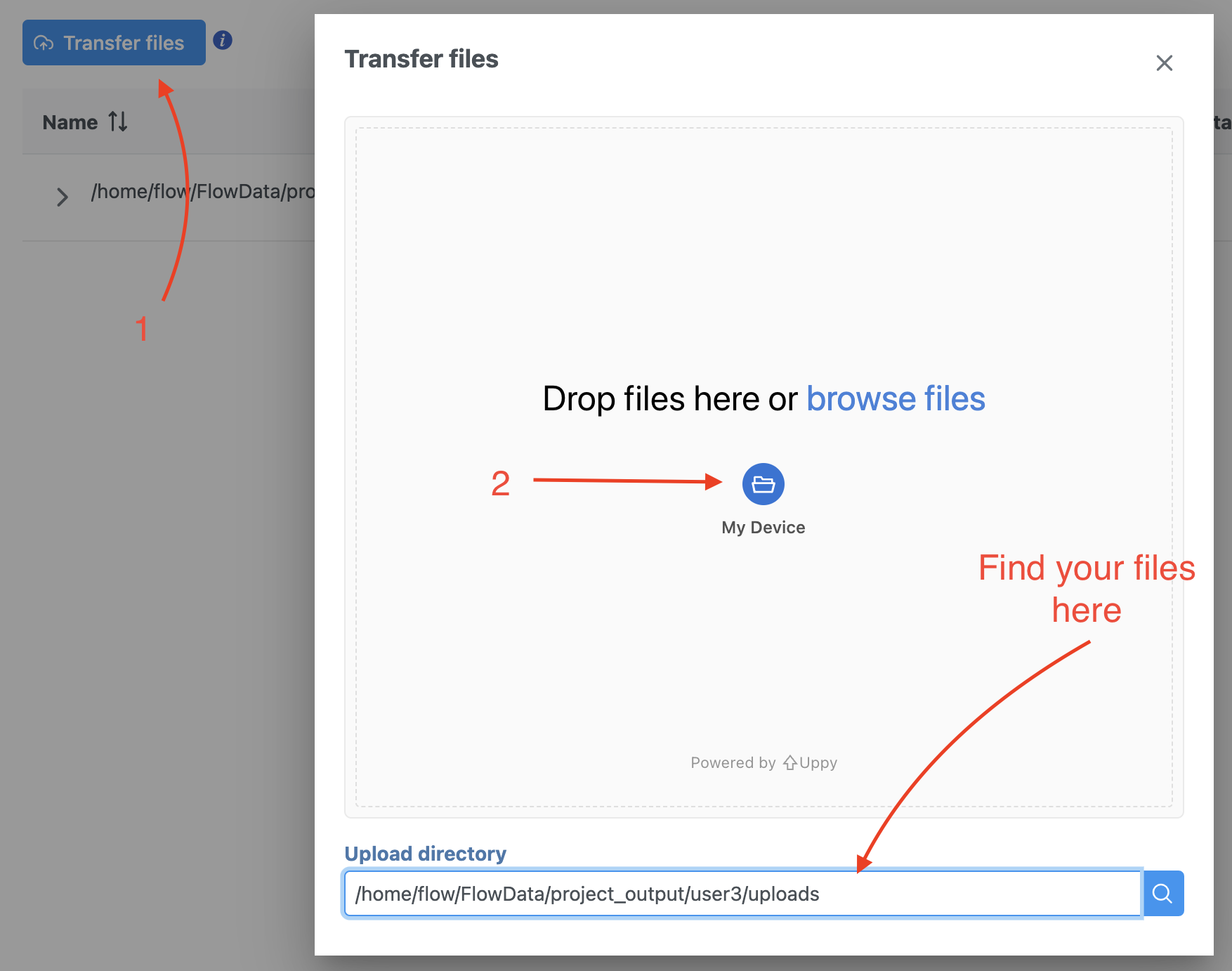 Image Added Image Added
|
Import the FASTQ files
To proceed, click the Import Add data button in the Data Analyses tab (Figure 2). Click the Automatically create samples from files buttonIn the Single cell > scATAC-Seq section select fastq and click Next. The file browser interface will open (Figure 3). Select the FASTQ files using the file browser interface and push the Create sample button Finish button to complete the task. Paired end reads will be automatically detected and multiple lanes for the same sample will be automatically combined into a single sample. We encourage users to include all the FASTQ files including the index files although they are optional.
When the FASTQ files have finished importing, the Unaligned reads data node will turn from transparent to opaqueappear in the Analyses tab.
| Numbered figure captions |
|---|
| SubtitleText | Data tab in Partek Flow. |
|---|
| AnchorName | Data tab |
|---|
|
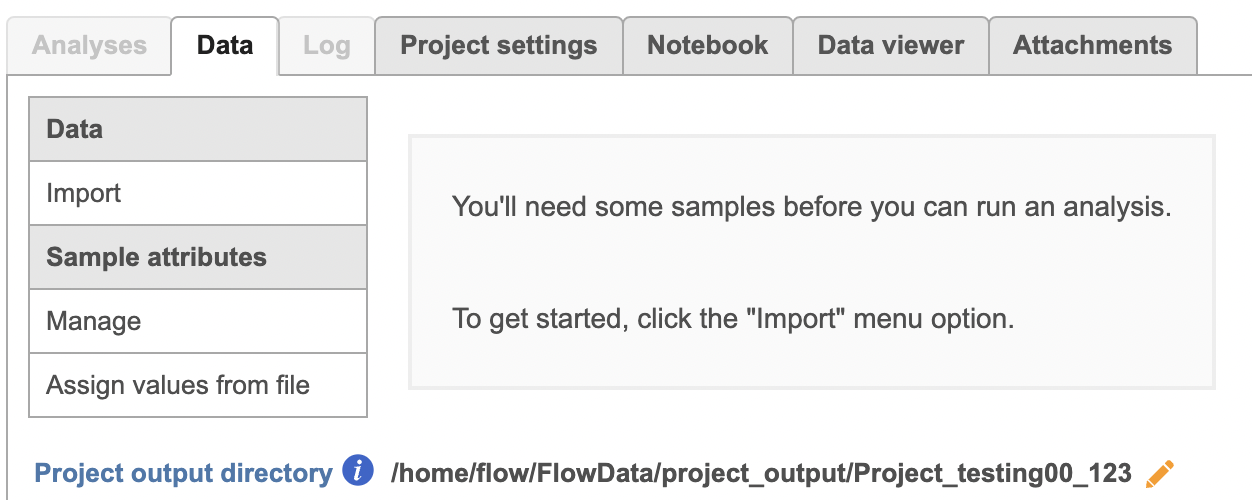 Image Removed Image Removed Image Added Image Added
|
| Numbered figure captions |
|---|
| SubtitleText | Input FASTQ files for scATAC-Seq data in Flow. |
|---|
| AnchorName | Input FASTQ files |
|---|
|
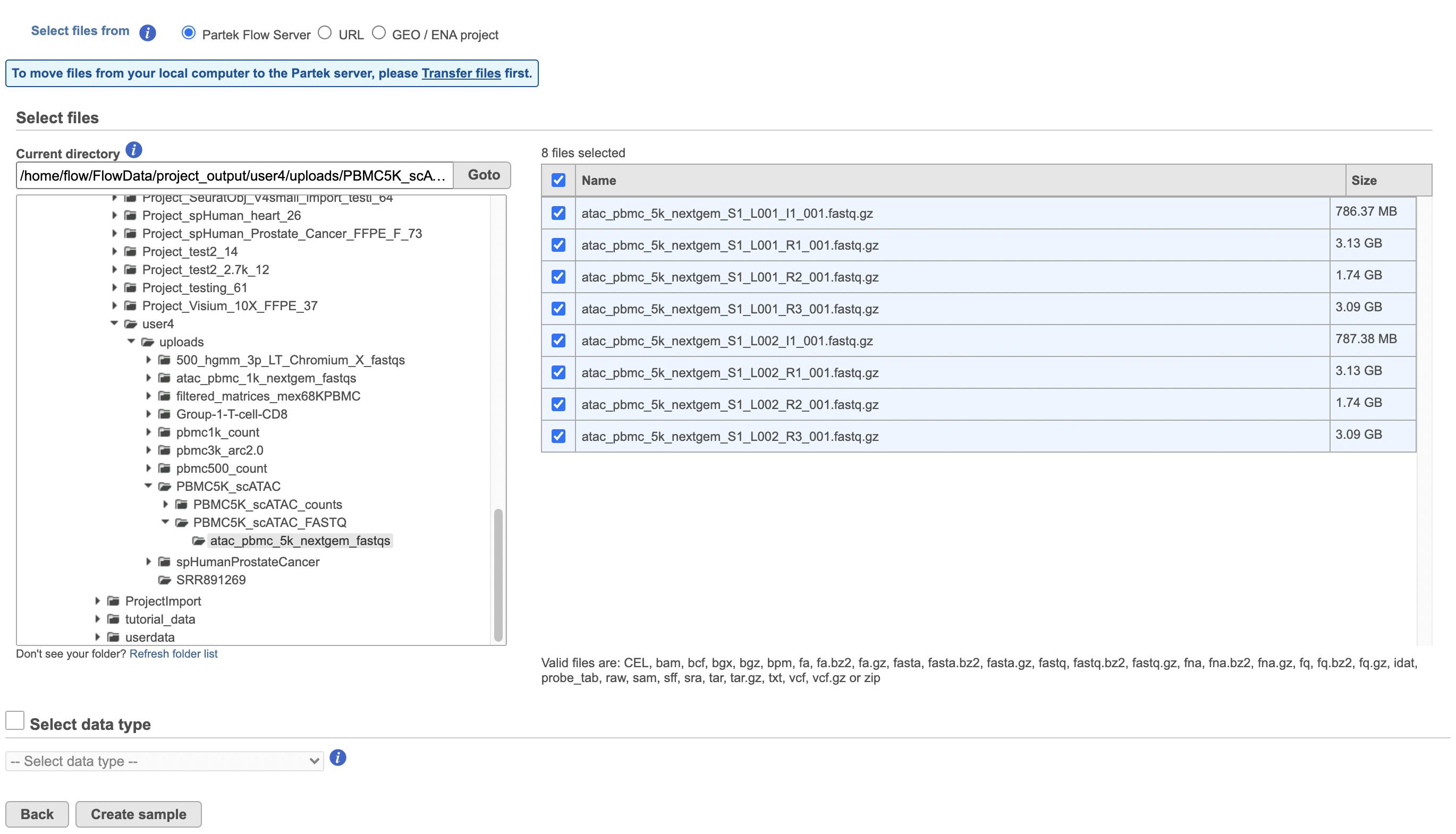 Image Removed Image Removed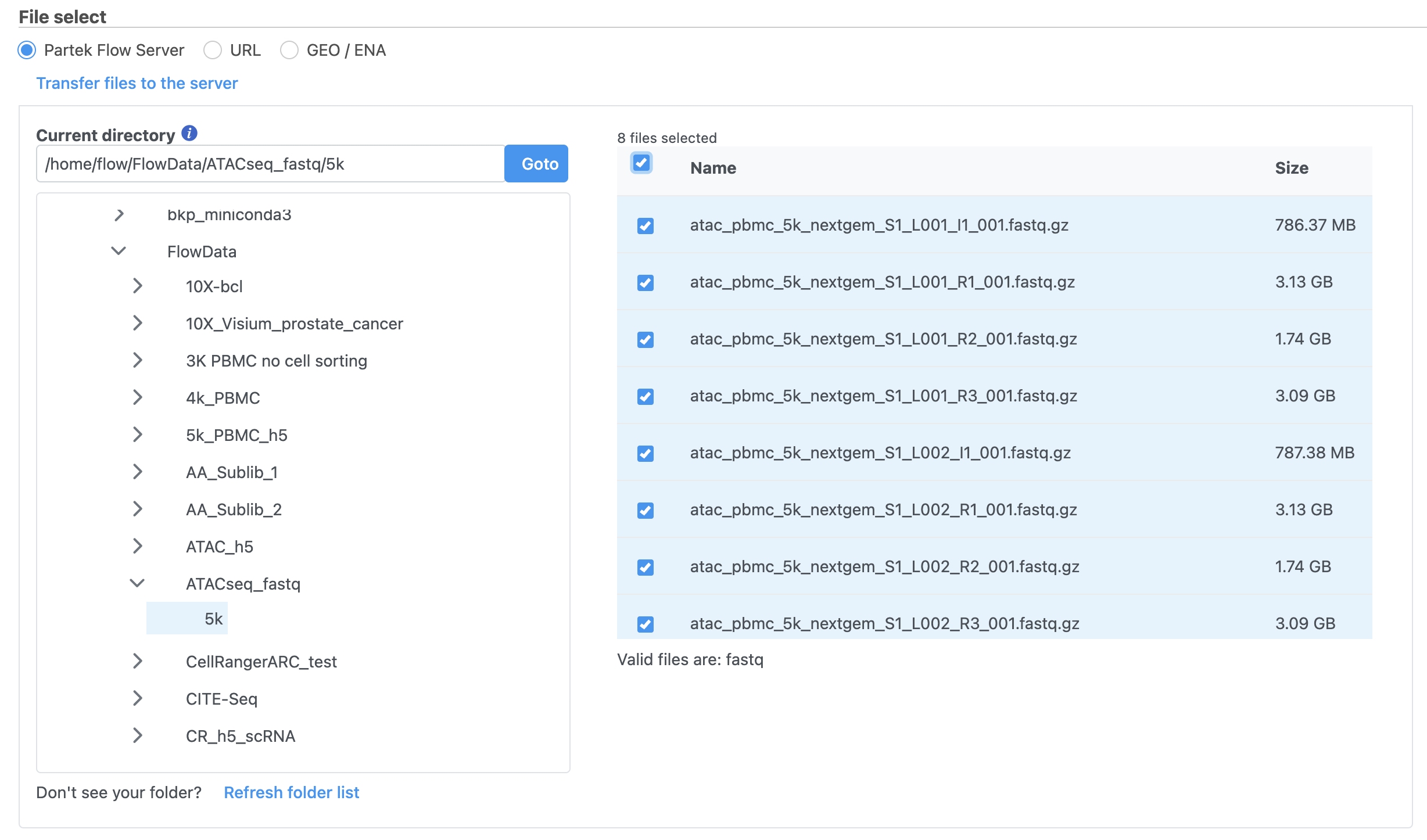 Image Added Image Added
|
Convert FASTQ to count
...
- Click the Unaligned reads data node
- Select Cell Ranger - ATAC in the 10x Genomics section in the task menu on the right
- Select Single cell ATAC in Assay type for ATAC-Seq data only
- Choose the proper Reference assembly for the data (you may have to create the reference)
- Press the Finish button to run the task with default settings (Figure 4)
| Numbered figure captions |
|---|
| SubtitleText | Convert FASTQ by Cell Ranger - ATAC task in Flow. |
|---|
| AnchorName | Cell Ranger - ATAC |
|---|
|
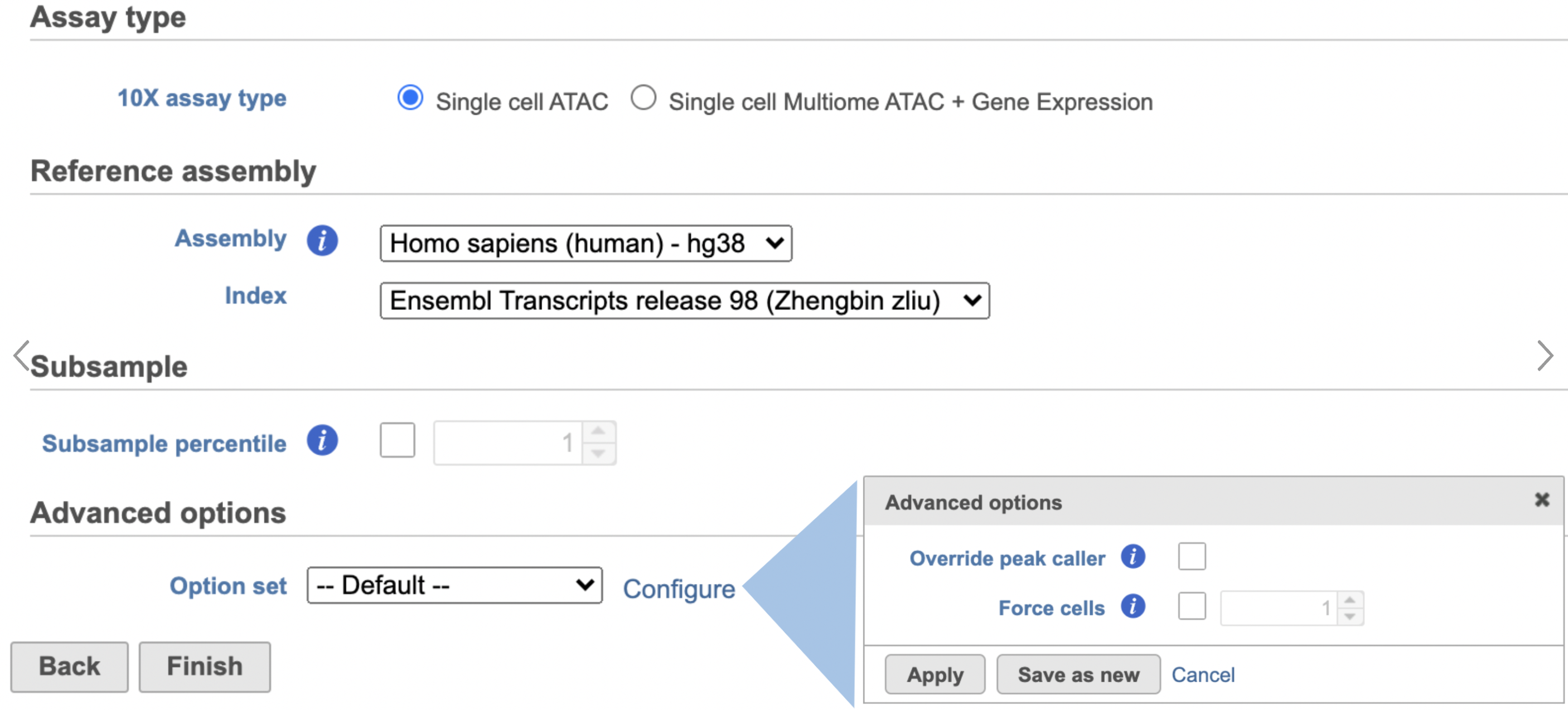 Image Removed Image Removed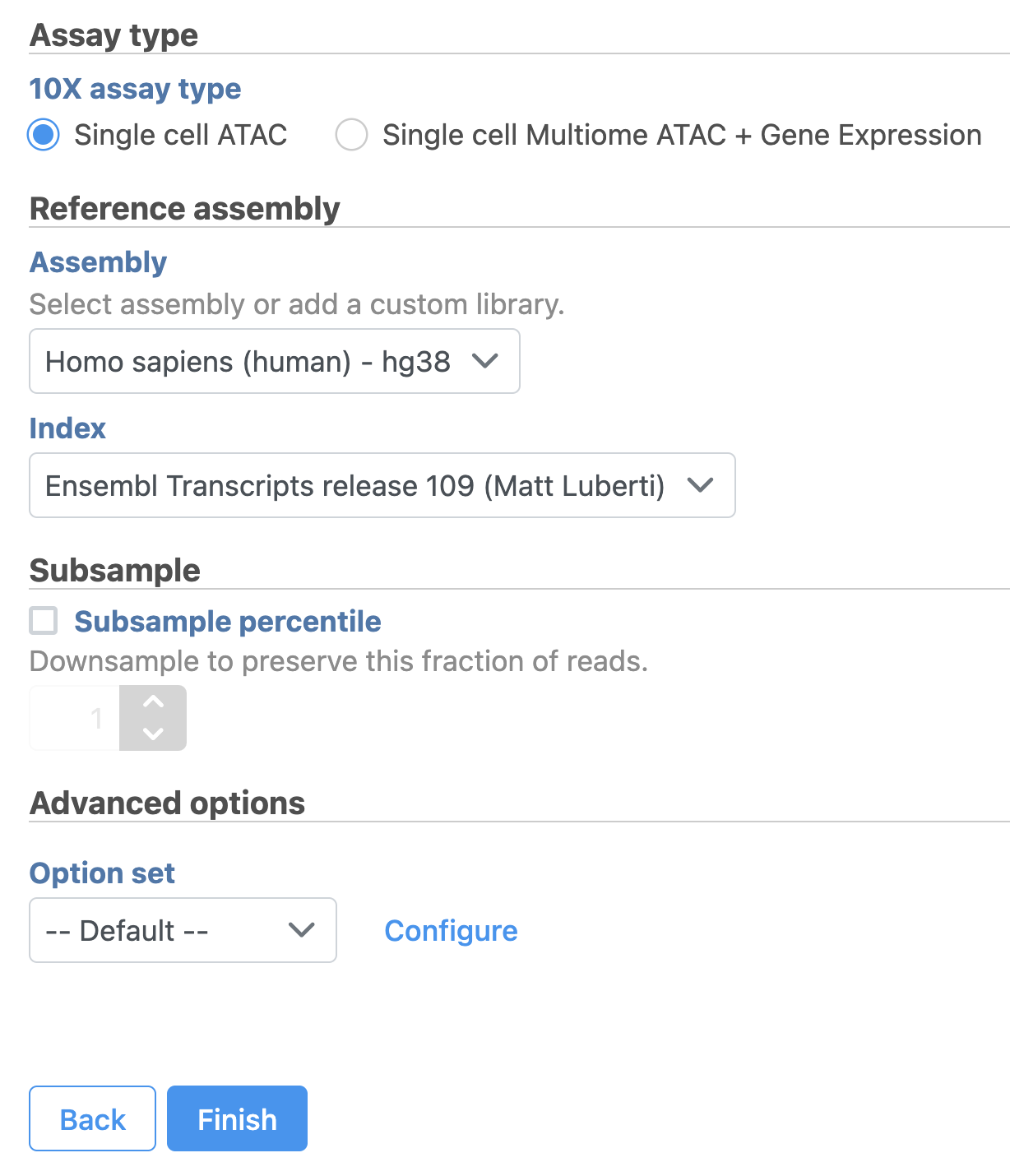 Image Added Image Added
|
To learn more about how to run Cell Ranger - ATAC task in Flow, please refer to our online documentation.
...
| Numbered figure captions |
|---|
| SubtitleText | Single cell QA/QC task for scATAC-Seq data in Flow. |
|---|
| AnchorName | QA/QC |
|---|
|
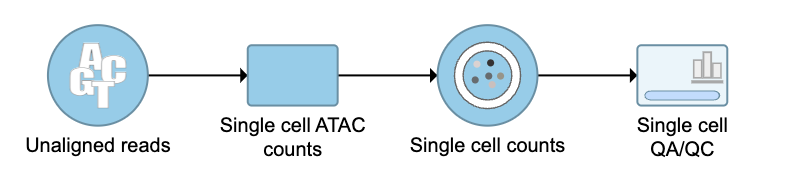 Image Removed Image Removed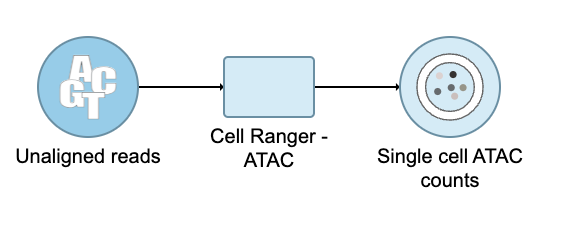 Image Added Image Added
|
QA/QC
An important step in analyzing single cell ATAC data is to filter out low quality cells. A few examples of low-quality cells are doublets, cells with a low TSS enrichment score, cells with a high proportion of reads mapping to the genomic blacklist regions, or cells with too few reads to be analyzed. Users are able to do this in Partek Flow using the Single cell QA/QC task.
...
| Numbered figure captions |
|---|
| SubtitleText | QA/QC task report for scATAC - Seq data in Flow. |
|---|
| AnchorName | QA/QC task report |
|---|
|
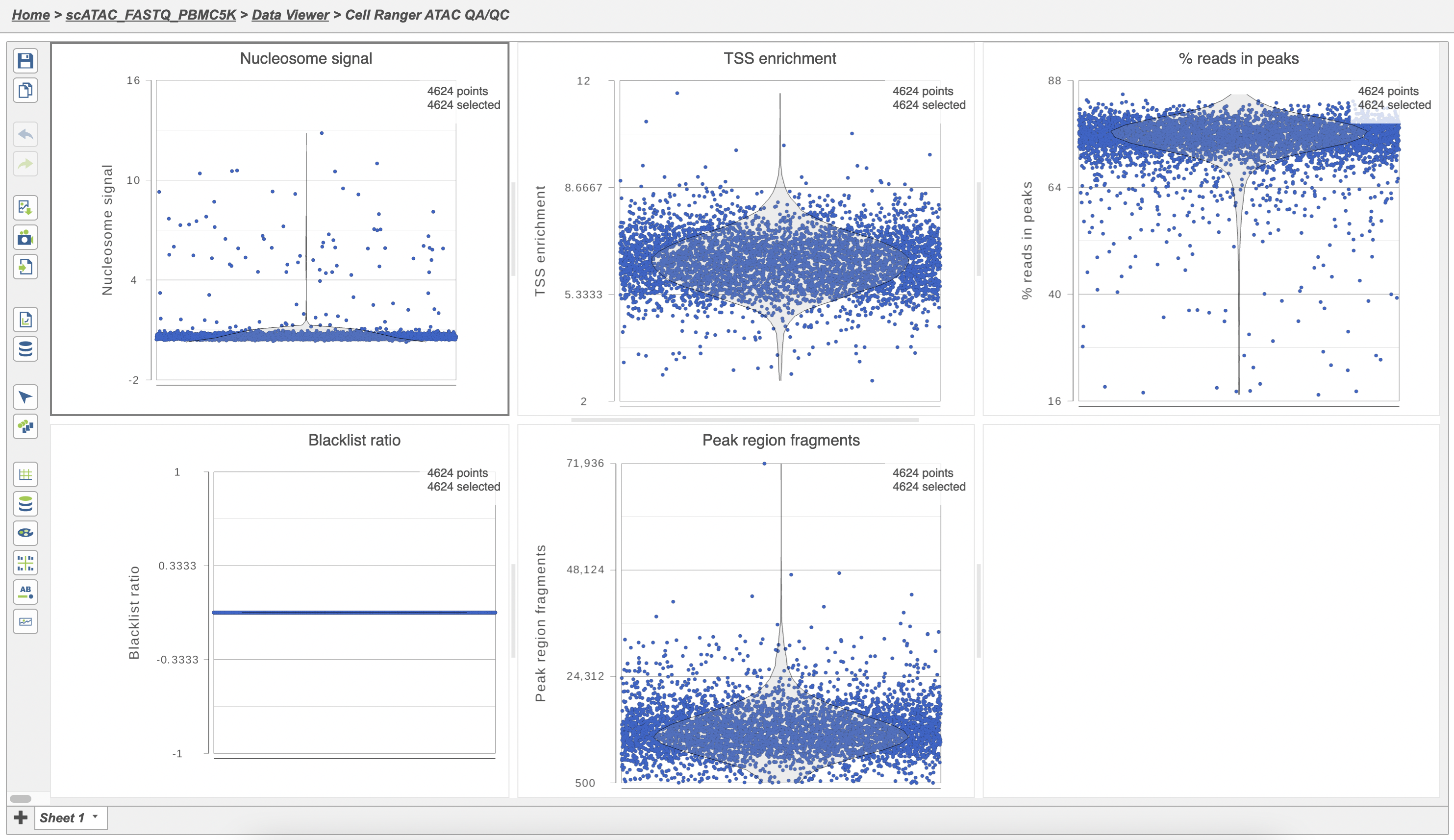 Image Removed Image Removed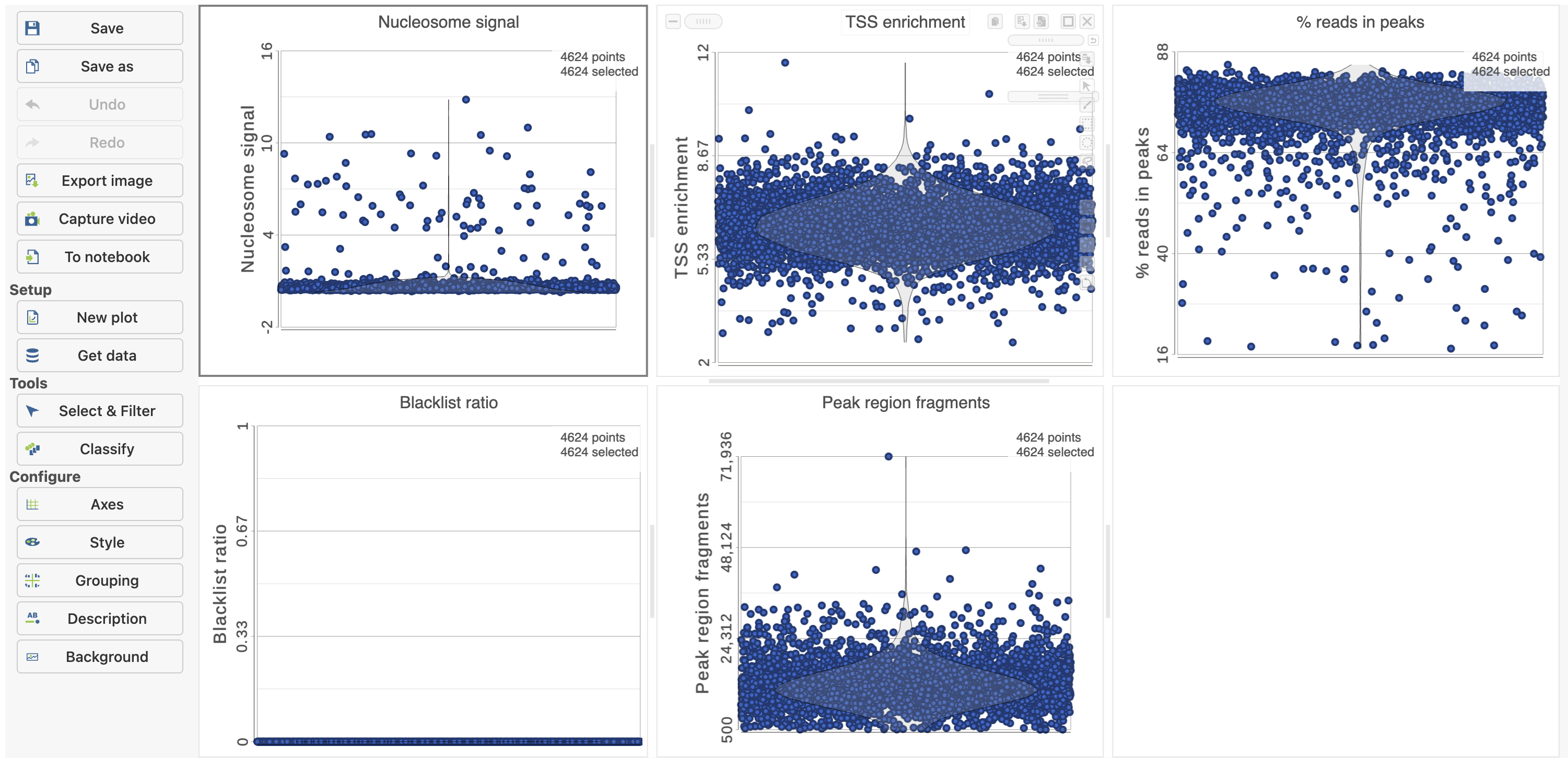 Image Added Image Added |
The Single cell QA/QC report includes interactive violin plots showing the value of every cell in the project on several quality measures (Figure 6).
...
To filter out low quality cells (Figure 7),
- Open the Select & Filter menu
- Set the filters on nucleosome signal < 4; Peak region fragment 500-30000; and % reads in peaks > 15% ; Blacklist ratio < 0.05leave the rest as they are
- Click the filter icon
 Image Addedand Apply observation filter to run the Filter cells task on the first Single cell ATAC counts data node, it generates a Filtered cells node
Image Addedand Apply observation filter to run the Filter cells task on the first Single cell ATAC counts data node, it generates a Filtered cells node - Click PCA from the drop-down list
| Numbered figure captions |
|---|
| SubtitleText | Filter low quality cells in Partek Flow. |
|---|
| AnchorName | Filter cells |
|---|
|
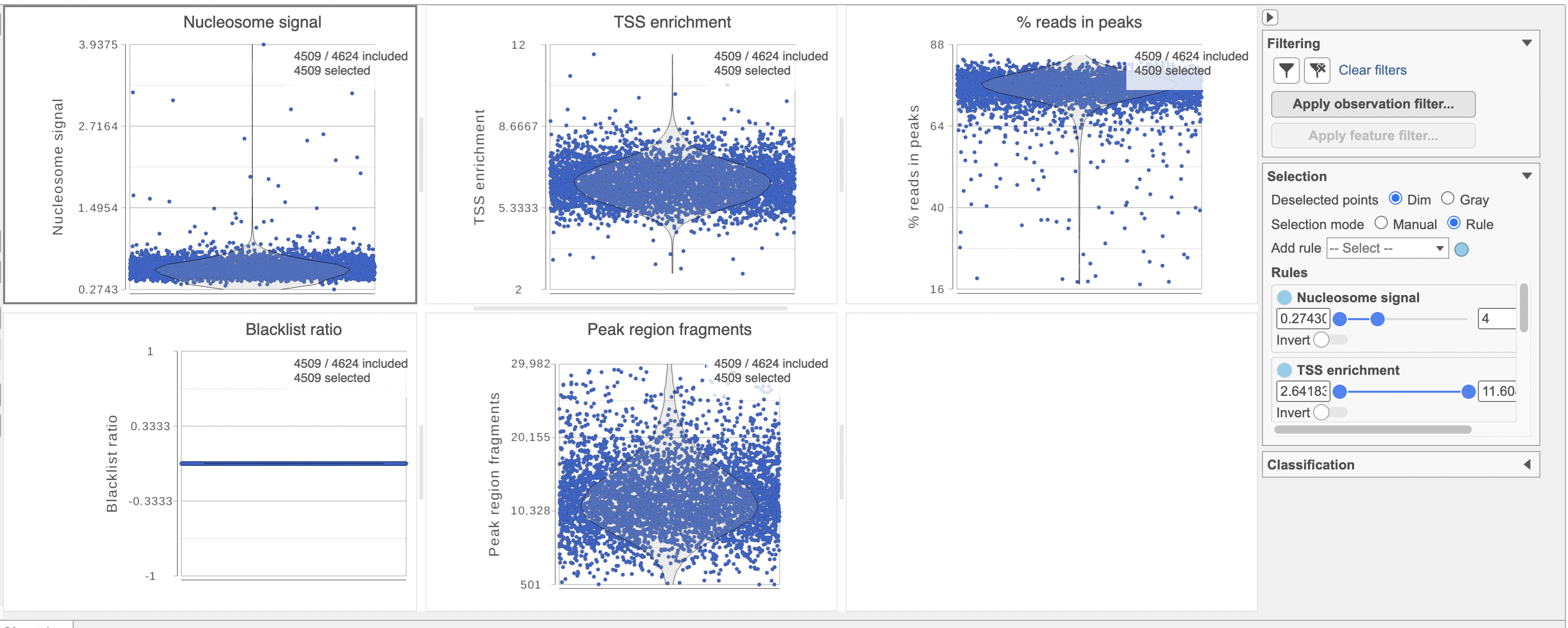 Image Removed Image Removed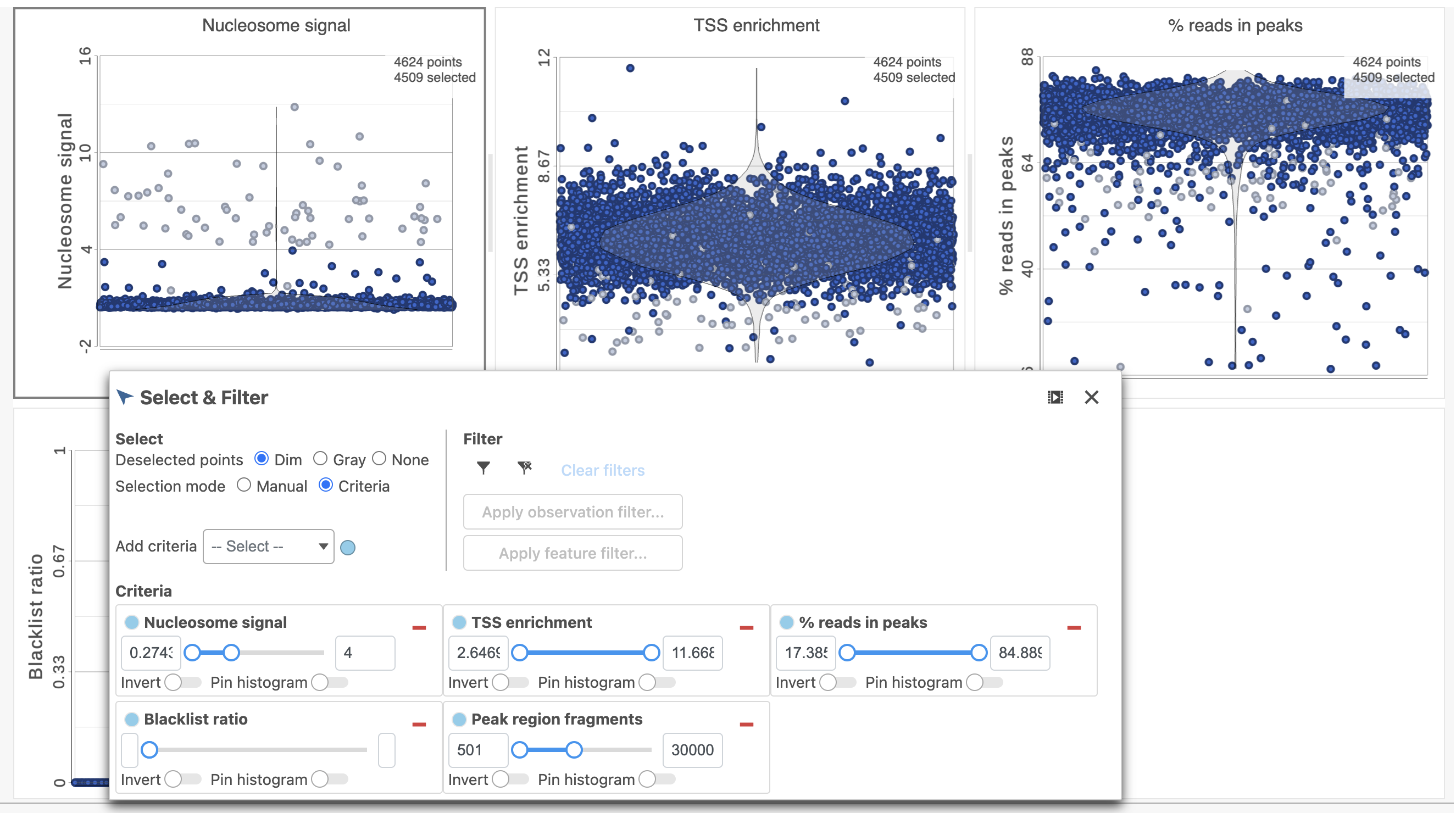 Image Added Image Added
|
Filter features
...
| Numbered figure captions |
|---|
| SubtitleText | Filter features in Partek Flow. |
|---|
| AnchorName | Filter features |
|---|
|
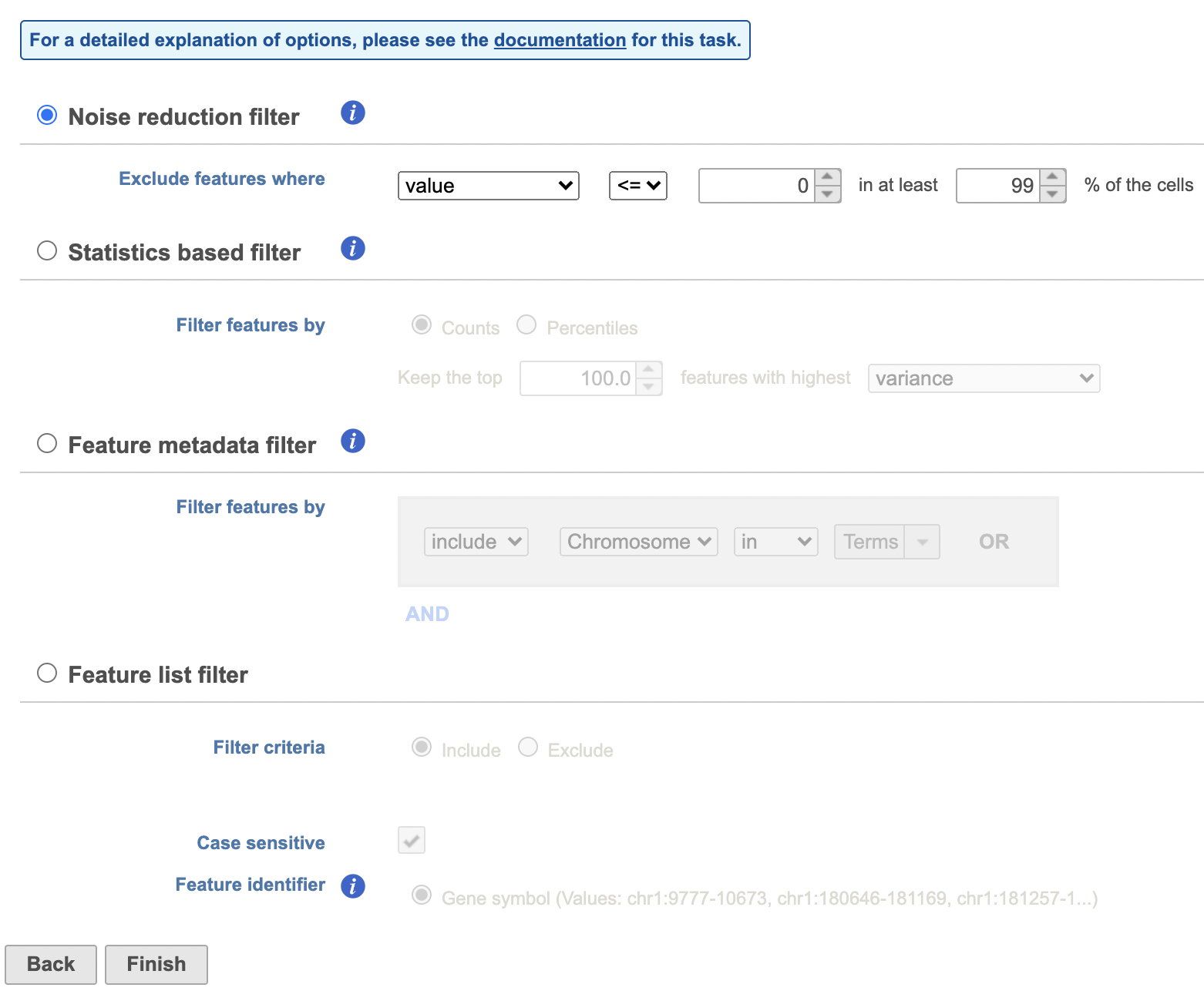 Image Removed Image Removed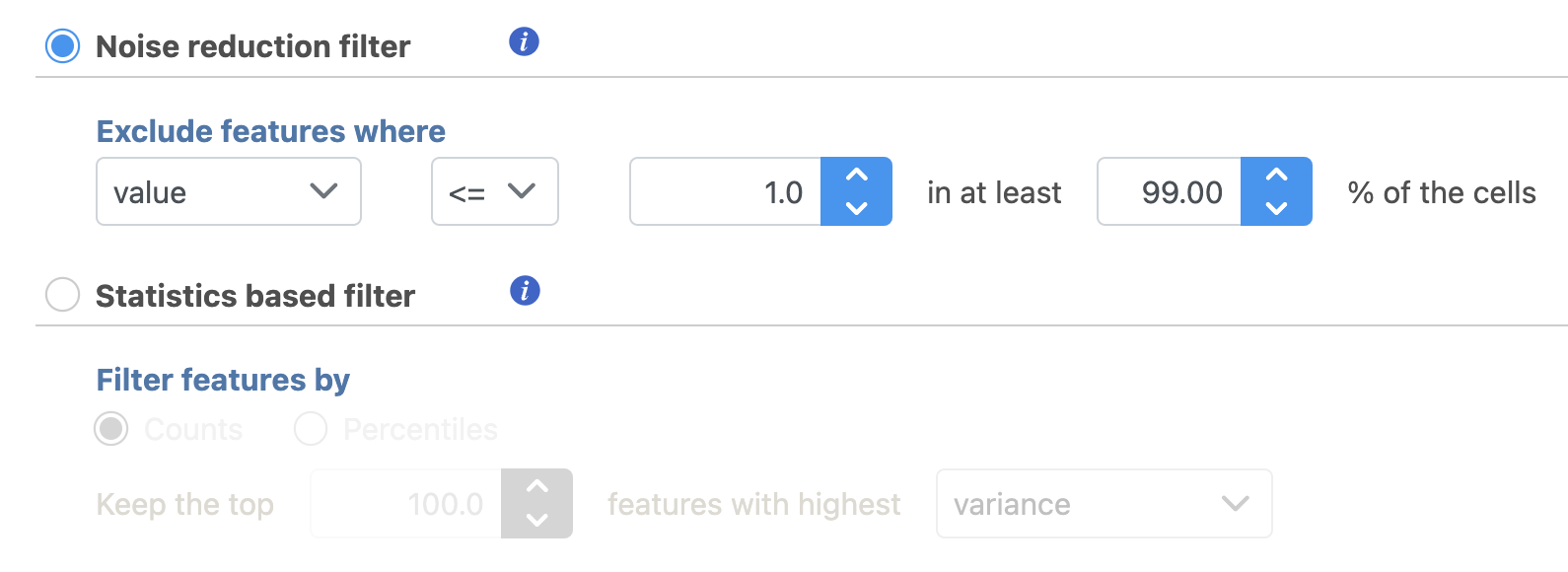 Image Added Image Added
|
Annotate regions
...
The input for Annotate peaks is a Peaks type data node.
- Click a Peaks the Filtered features data node
- Click the Peak analysis section in the toolbox
- Click Annotate regions
- Set the Genomic overlaps parameter
...
| Numbered figure captions |
|---|
| SubtitleText | Annotate regions in Partek Flow. |
|---|
| AnchorName | Annotate regions |
|---|
|
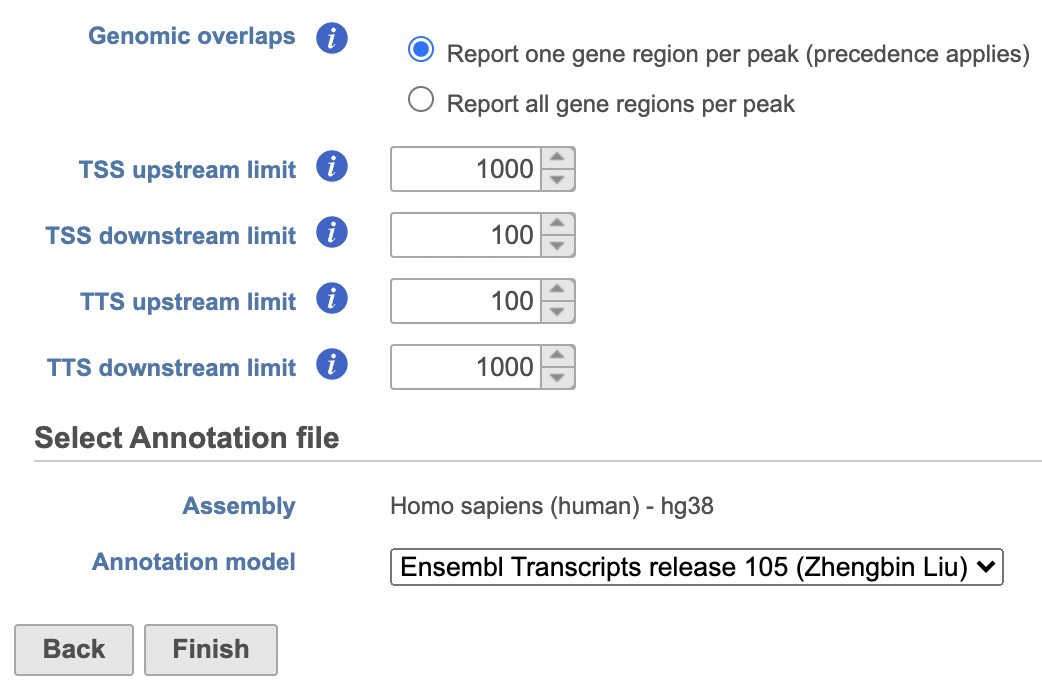 Image Removed Image Removed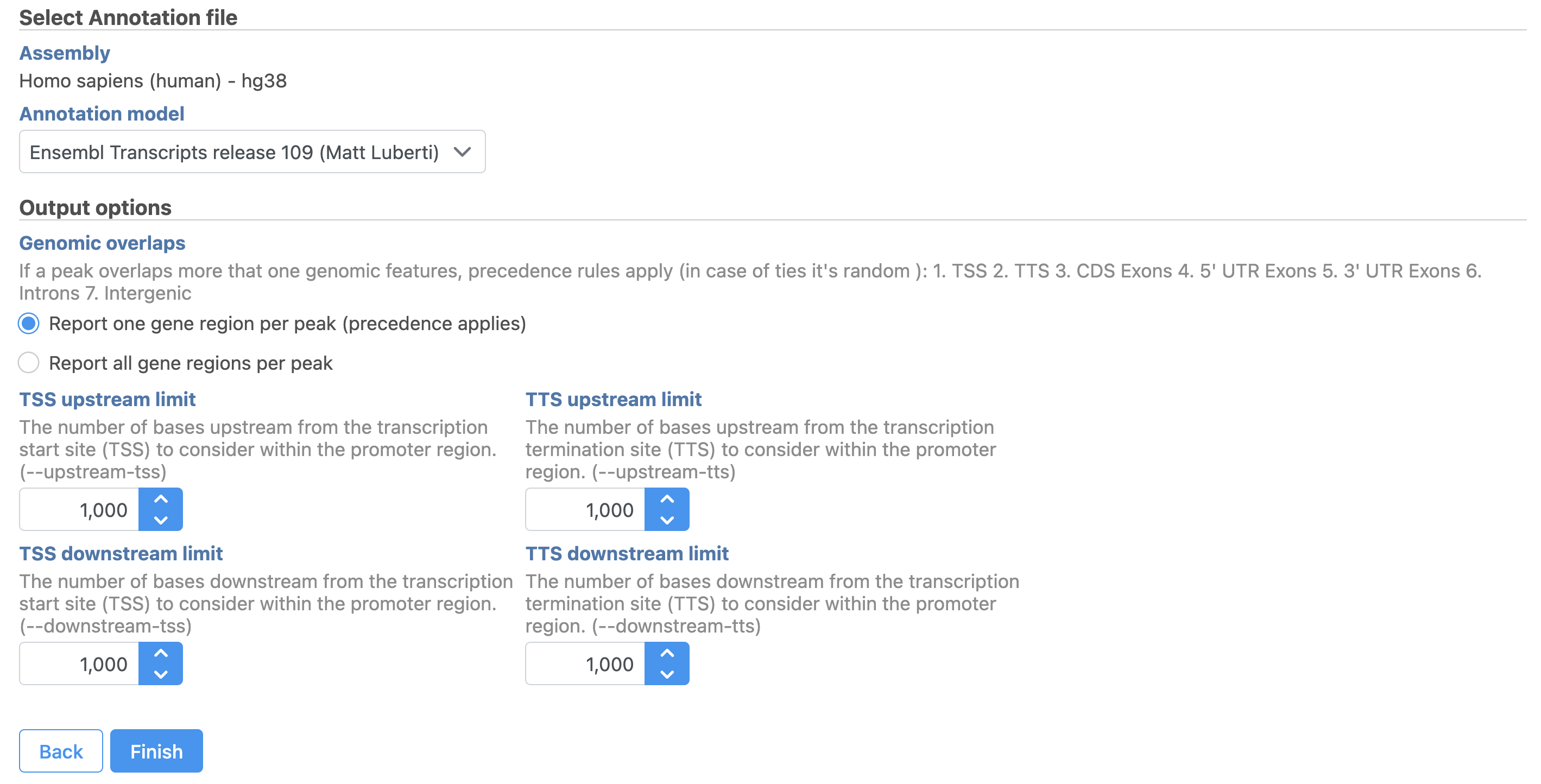 Image Added Image Added
|
Users are able to define the transcription start site (TSS) and transcription termination site (TTS) limit in the unit of bp.
...
| Numbered figure captions |
|---|
| SubtitleText | TF-IDF normalization for scATAC-Seq in Flow. |
|---|
| AnchorName | Normalization |
|---|
|
 Image Removed Image Removed Image Added Image Added
|
To run TF-IDF normalization,
- Click a single Single cell counts data node, in this case the Annotated regions node
- Click the Normalization and scaling section in the toolbox
- Click TF-IDF normalization
...
| Numbered figure captions |
|---|
| SubtitleText | SVD task configuration dialog in Partek Flow. |
|---|
| AnchorName | SVD |
|---|
|
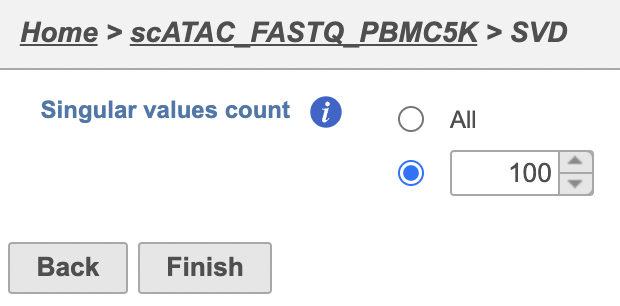 Image Removed Image Removed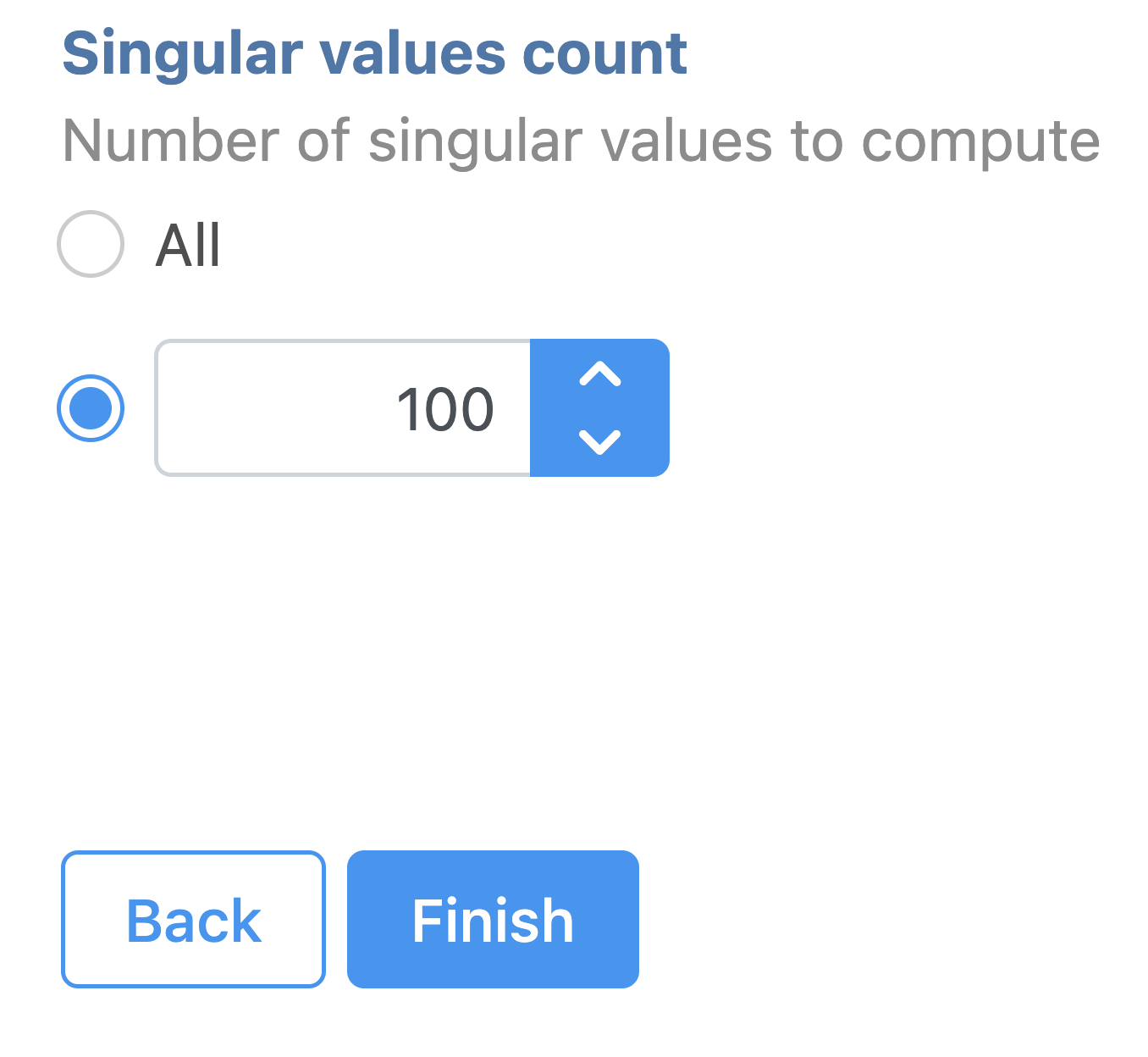 Image Added Image Added
|
Graph-based clustering
...
| Numbered figure captions |
|---|
| SubtitleText | Configure Graph-based clustering in Flow. |
|---|
| AnchorName | Configure Graph-based clustering |
|---|
|
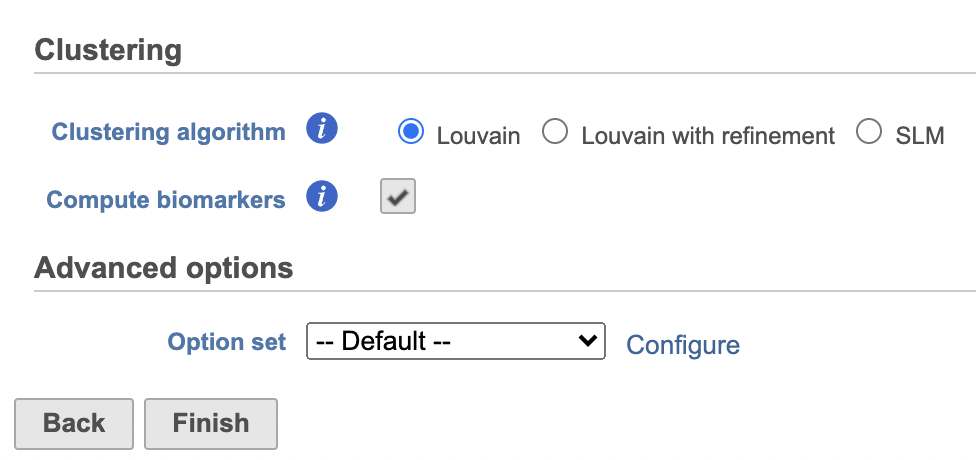 Image Removed Image Removed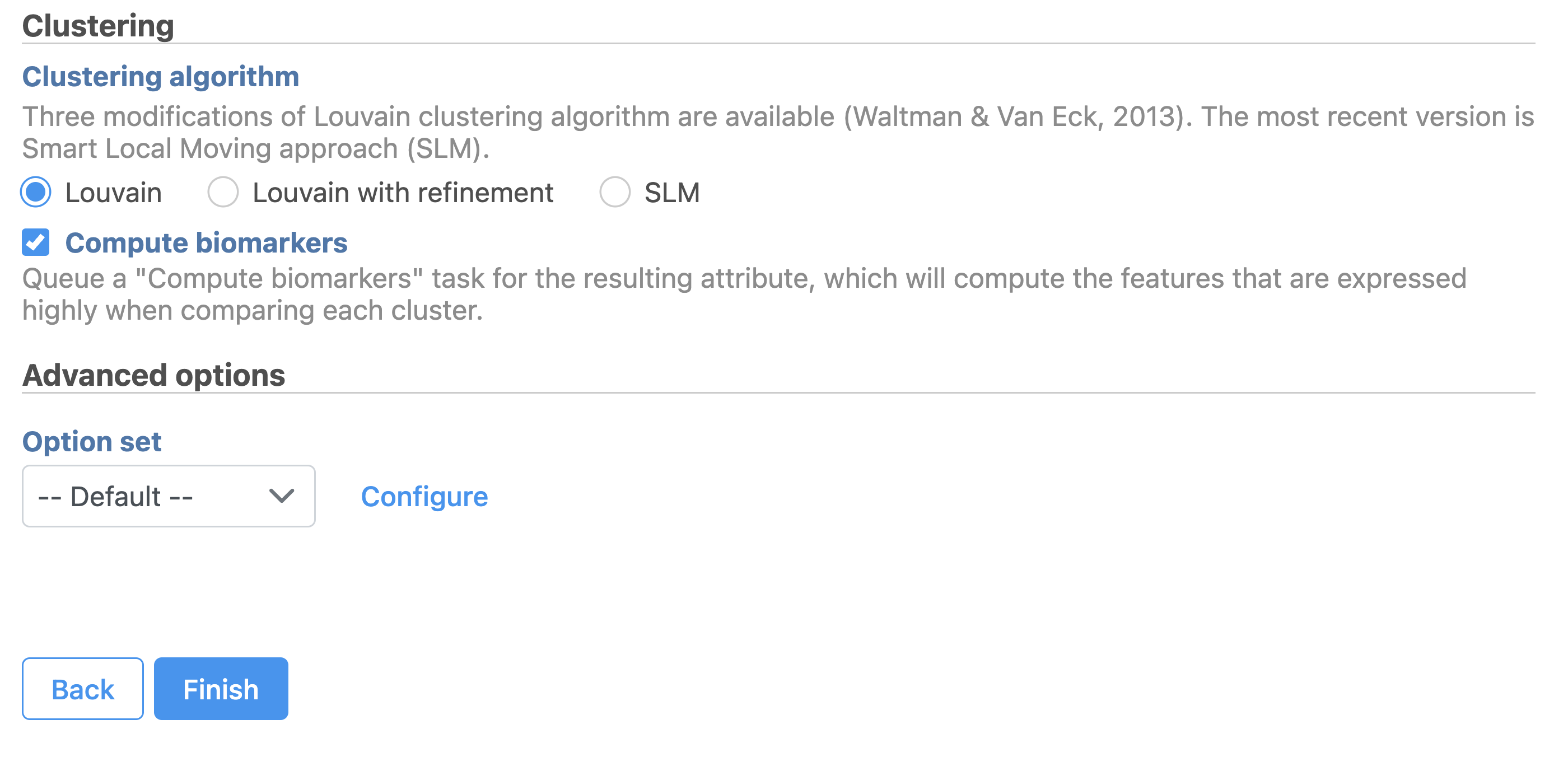 Image Added Image Added
|
A new Graph-based clusters data and a Biomarkers data node will be generated.
...
| Numbered figure captions |
|---|
| SubtitleText | Graph-based clustering results in Flow. |
|---|
| AnchorName | Graph-based clustering results |
|---|
|
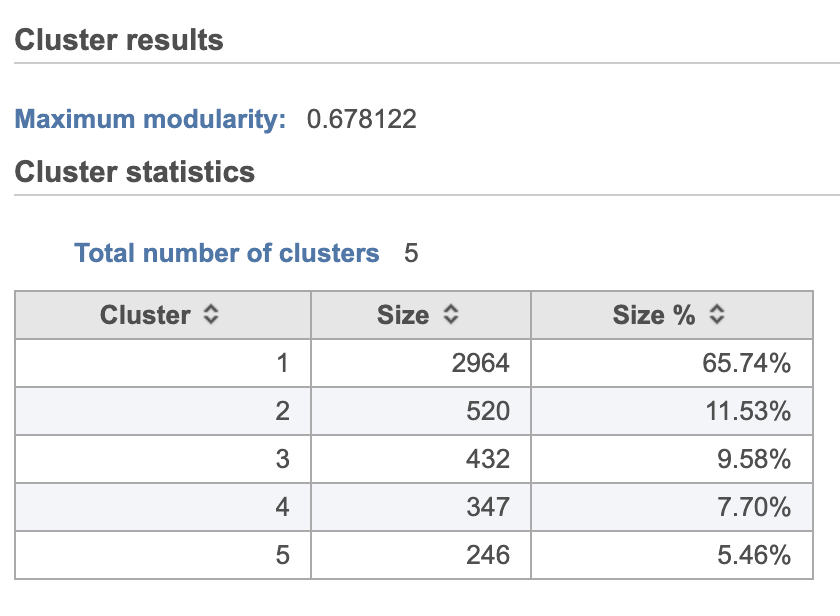 Image Removed Image Removed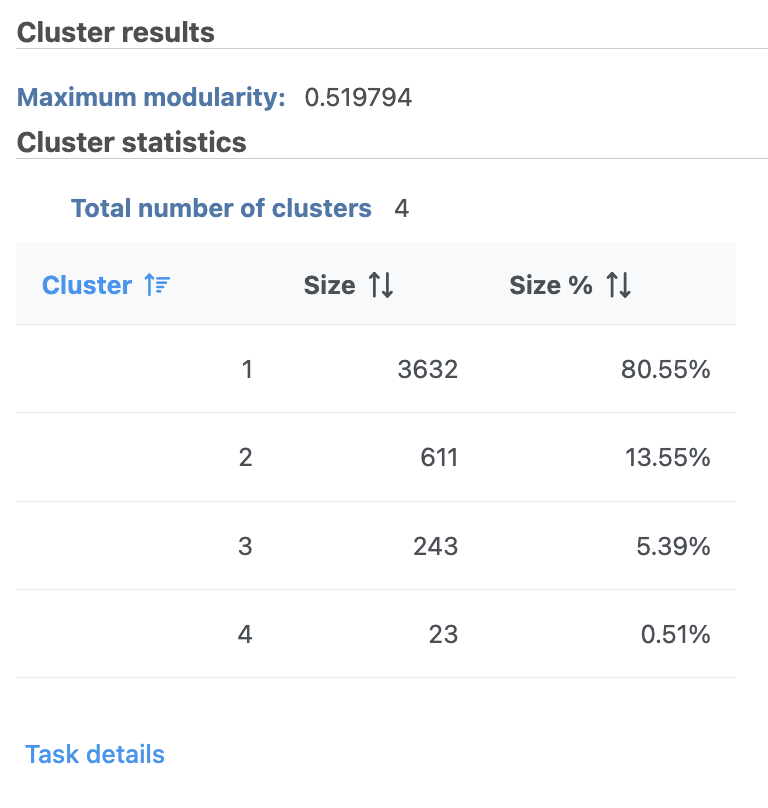 Image Added Image Added
|
| Numbered figure captions |
|---|
| SubtitleText | Computer biomarkers results in Flow. |
|---|
| AnchorName | Computer biomarkers results |
|---|
|
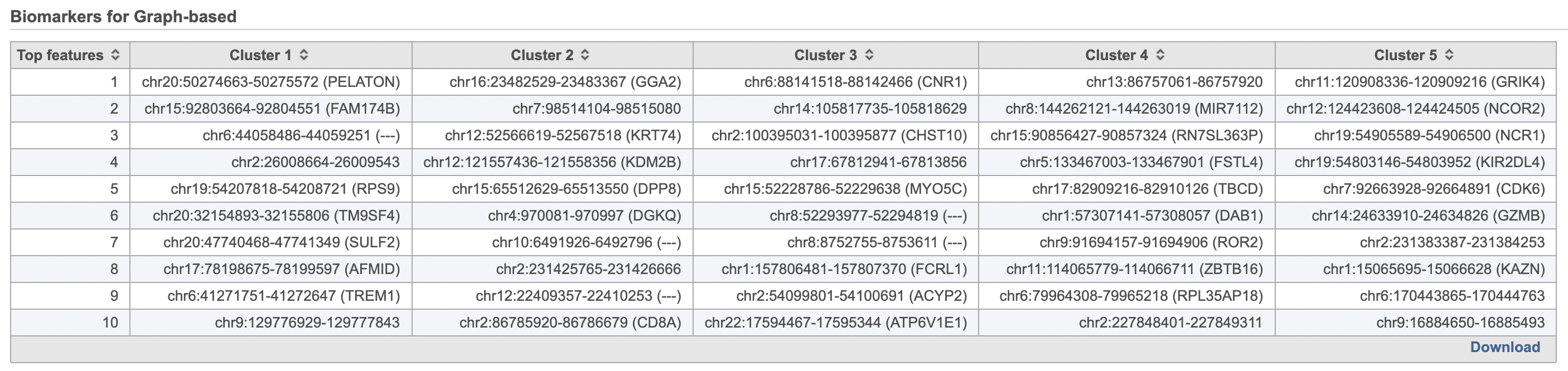 Image Removed Image Removed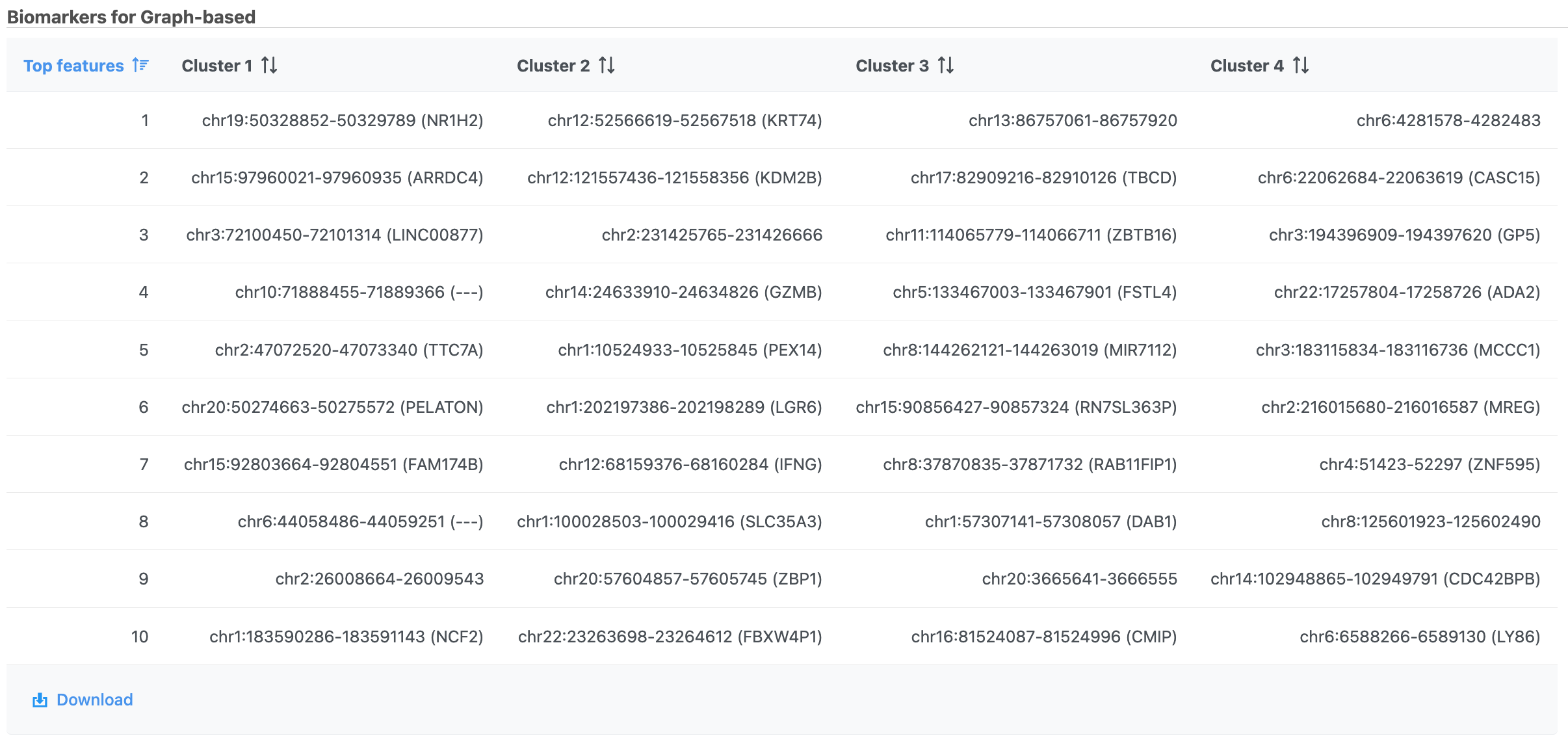 Image Added Image Added
|
UMAP
Similar to t-SNE, Uniform Manifold Approximation and Projection (UMAP) is a dimensional reduction technique. UMAP aims to preserve the essential high-dimensional structure and present it in a low-dimensional representation. UMAP is particularly useful for visually identifying groups of similar samples or cells in large high-dimensional data sets.
...
| Numbered figure captions |
|---|
| SubtitleText | UMAP configuration in Partek Flow. |
|---|
| AnchorName | UMAP configuration |
|---|
|
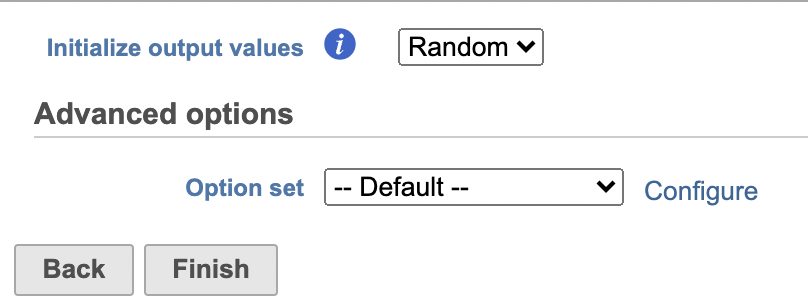 Image Removed Image Removed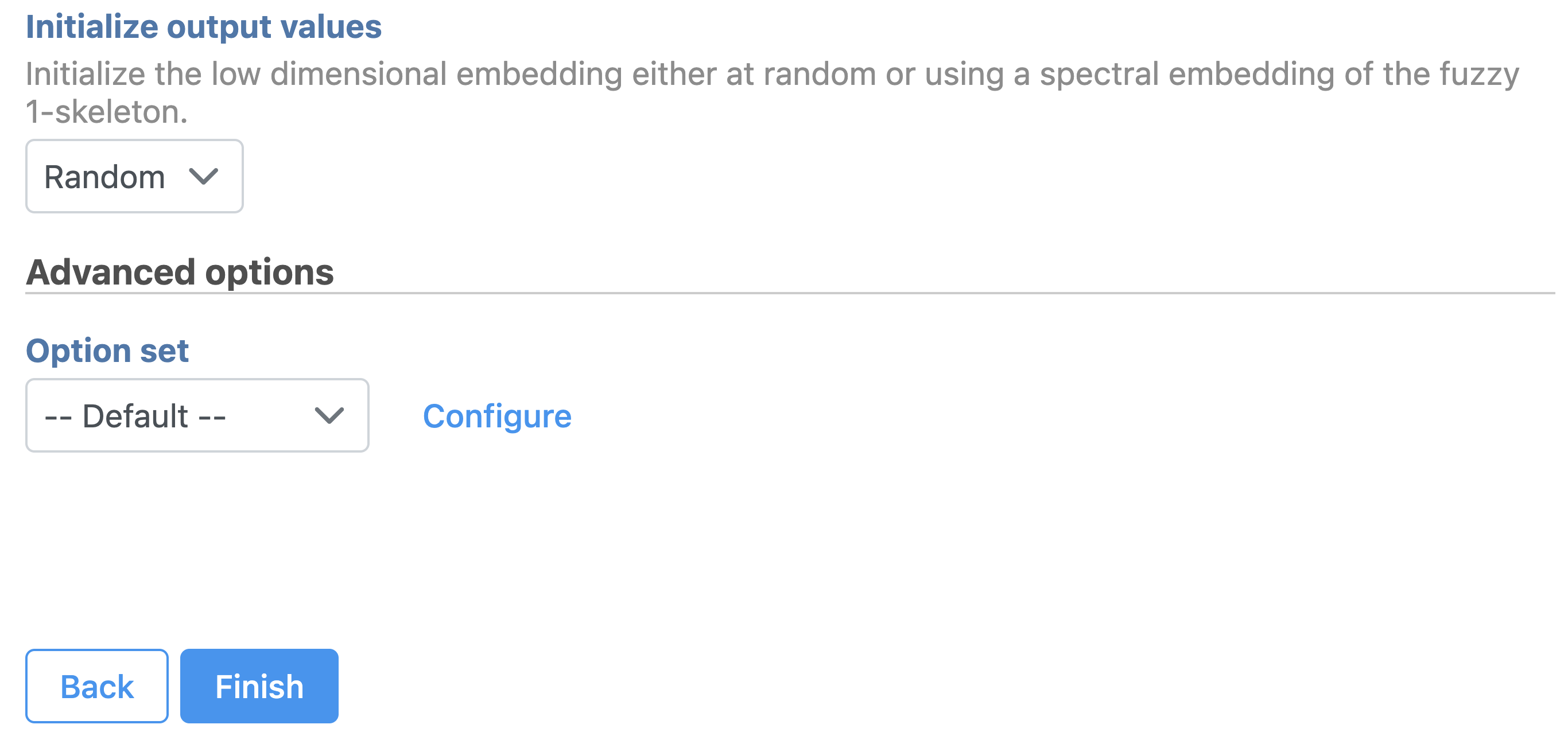 Image Added Image Added
|
Promoter sum matrix
...
| Numbered figure captions |
|---|
| SubtitleText | Promoter sum matrix in Flow. |
|---|
| AnchorName | Promoter sum matrix |
|---|
|
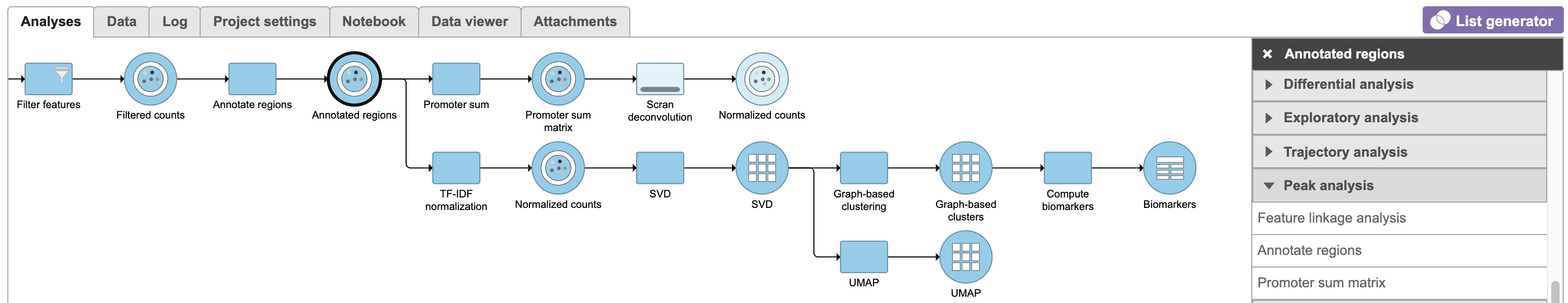 Image Removed Image Removed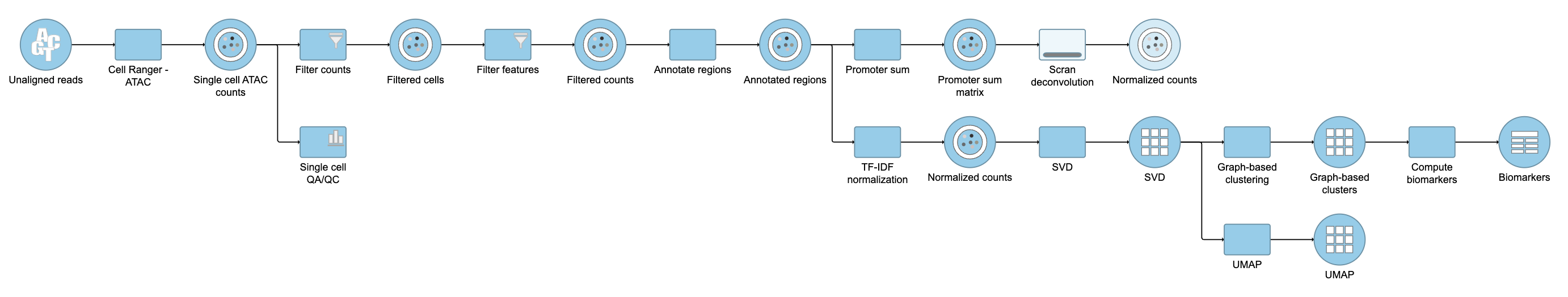 Image Added Image Added
|
Classifying cells
...
- Make sure the right data source has been selected. For scATAC-seq data, it shall be the normalized counts of promoter sum values in most cases (Figure 17)
- Set Color by in the Style configuration to the normalized counts node
- Type MS4A1 in the search box and select it. Rotate the 3D plot if you need to see this cluster more clearly.
- Click Click
 Image Added to activate Lasso mode
Image Added to activate Lasso mode - Draw a lasso around the cluster of MS4A1-expressing cells
- Click Classify selection under Tools in the left panel
- Type B cells for the Name
- Click Save (Figure 18)
...
| Numbered figure captions |
|---|
| SubtitleText | Select the data source in Data Viewer. |
|---|
| AnchorName | Select the data node |
|---|
|
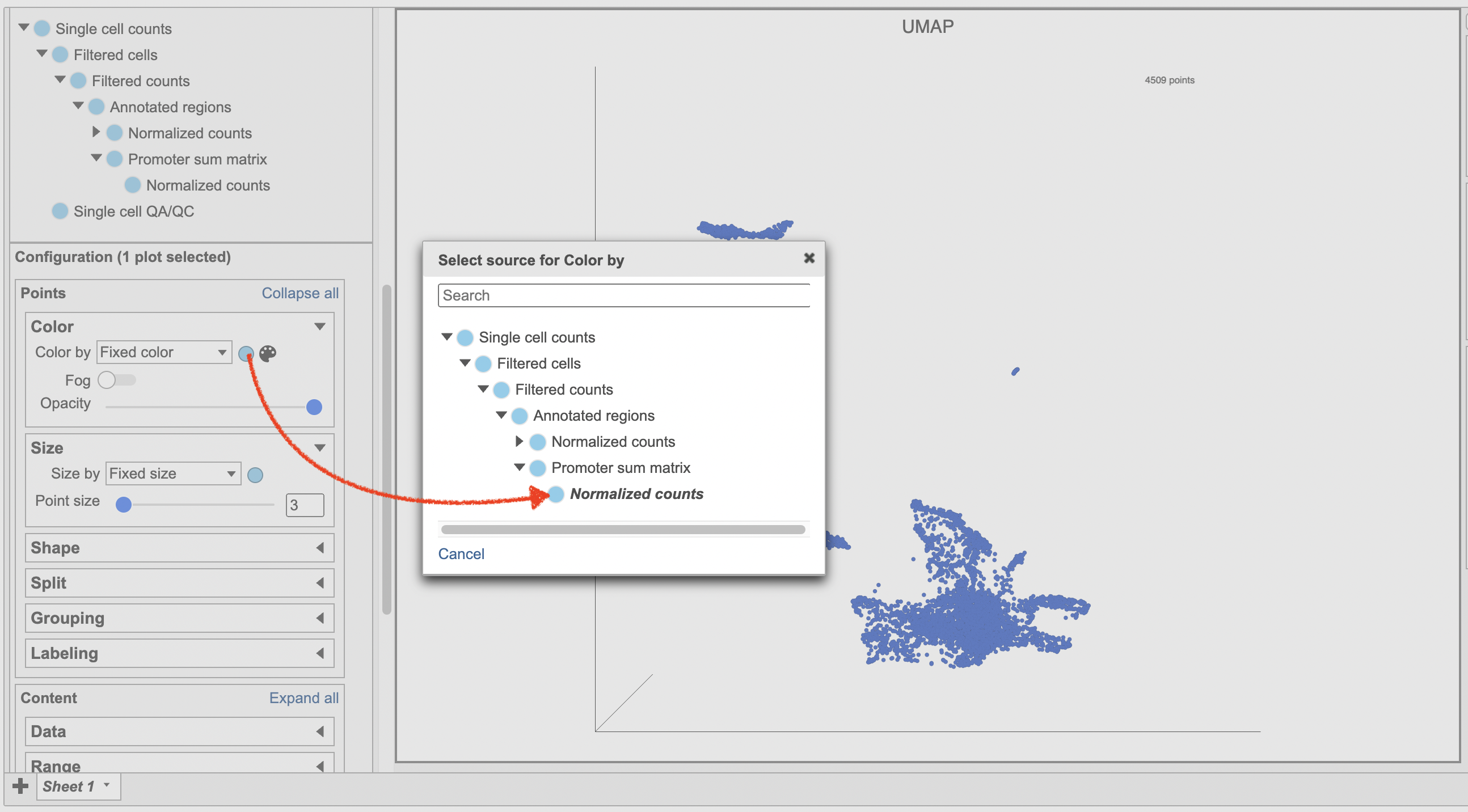 Image Removed Image Removed Image Added Image Added
|
| Numbered figure captions |
|---|
| SubtitleText | Color cells in UMAP by MS4A1 in Flow. |
|---|
| AnchorName | Coloring by MS4A1 |
|---|
|
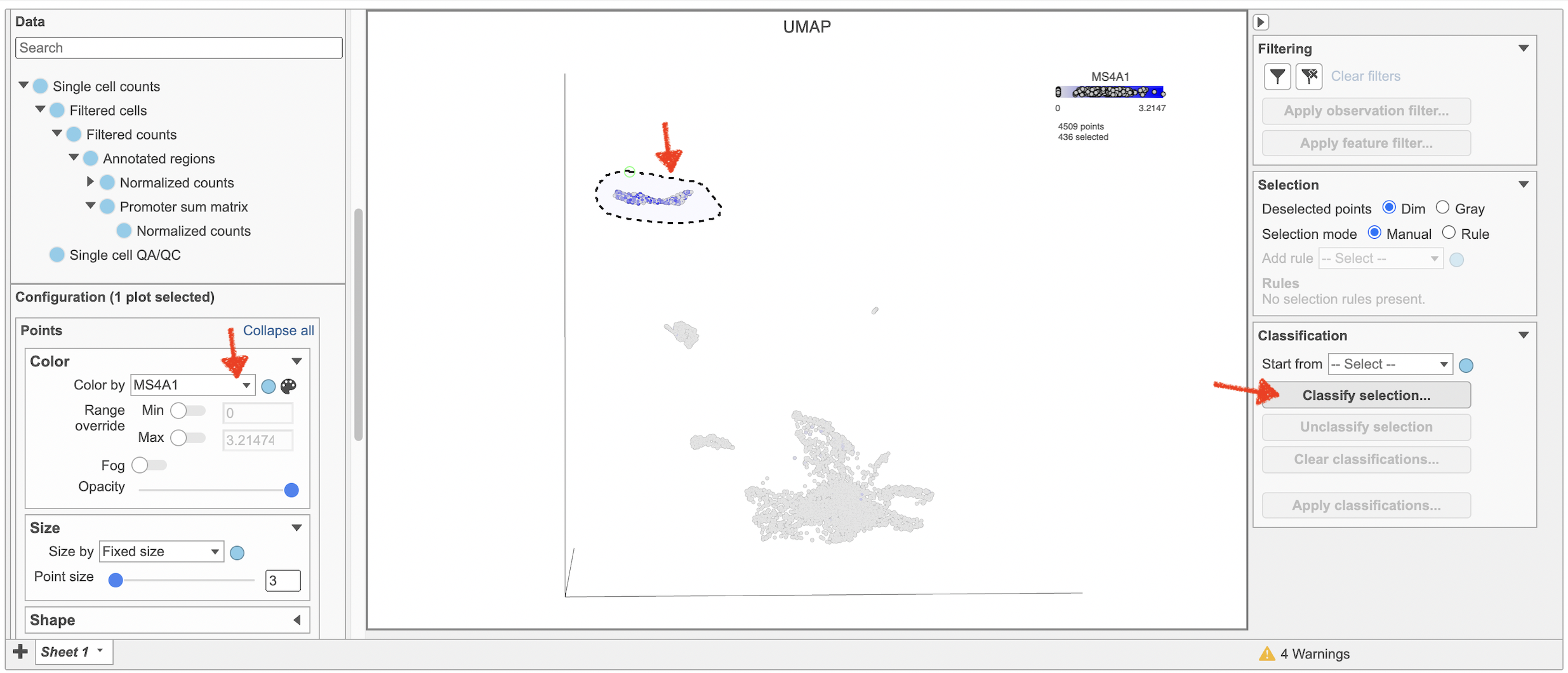 Image Removed Image Removed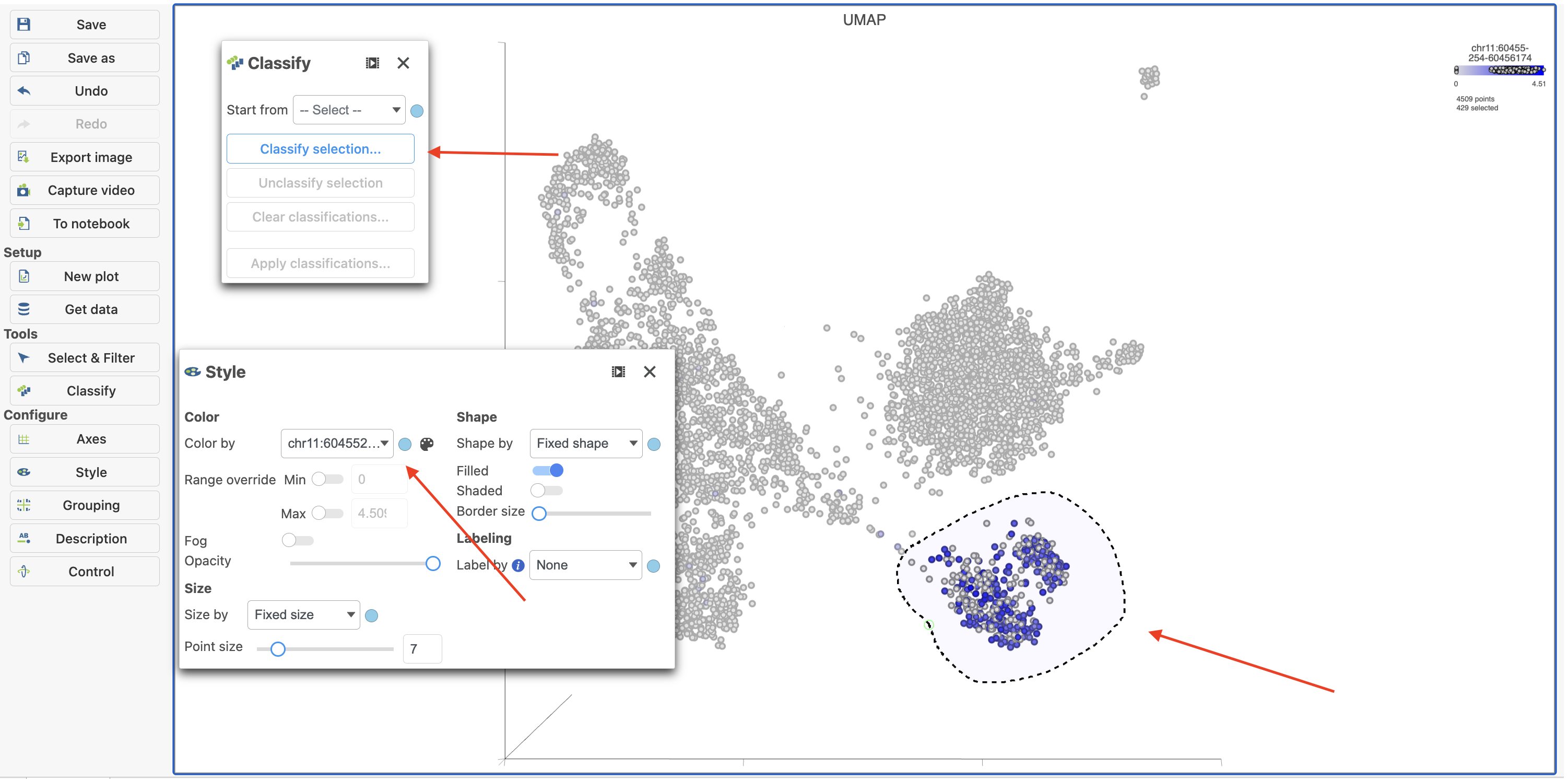 Image Added Image Added
|
Differential analysis
...
| Numbered figure captions |
|---|
| SubtitleText | Hurdle model for differential analysis in Flow. |
|---|
| AnchorName | Hurdle model |
|---|
|
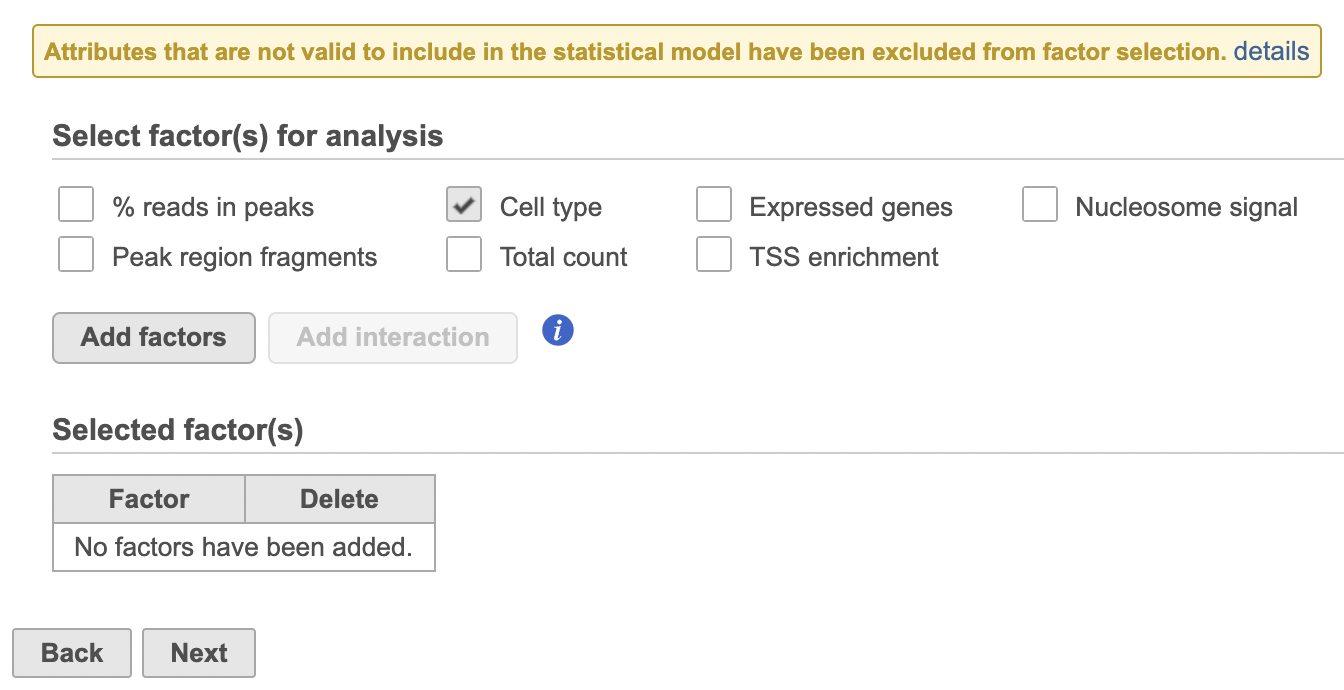 Image Removed Image Removed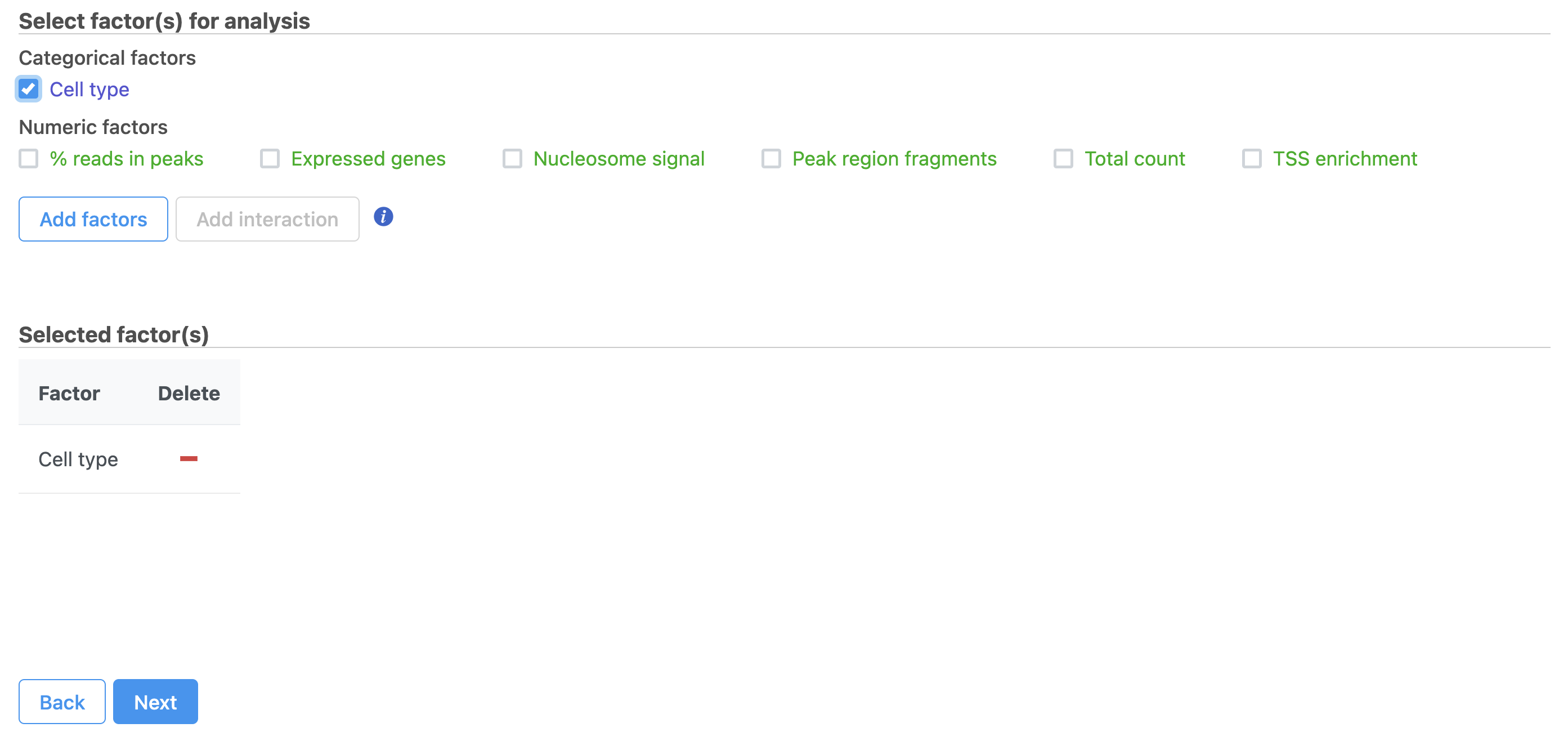 Image Added Image Added
|
- Click Next
- Define comparisons between factor or interaction levels (Figure 20)
- Click Add comparison to add the comparison to the Comparisons table.
- Click Finish to run the statistical test as default
| Numbered figure captions |
|---|
| SubtitleText | Define comparisons in Hurdle model. |
|---|
| AnchorName | Define comparisons |
|---|
|
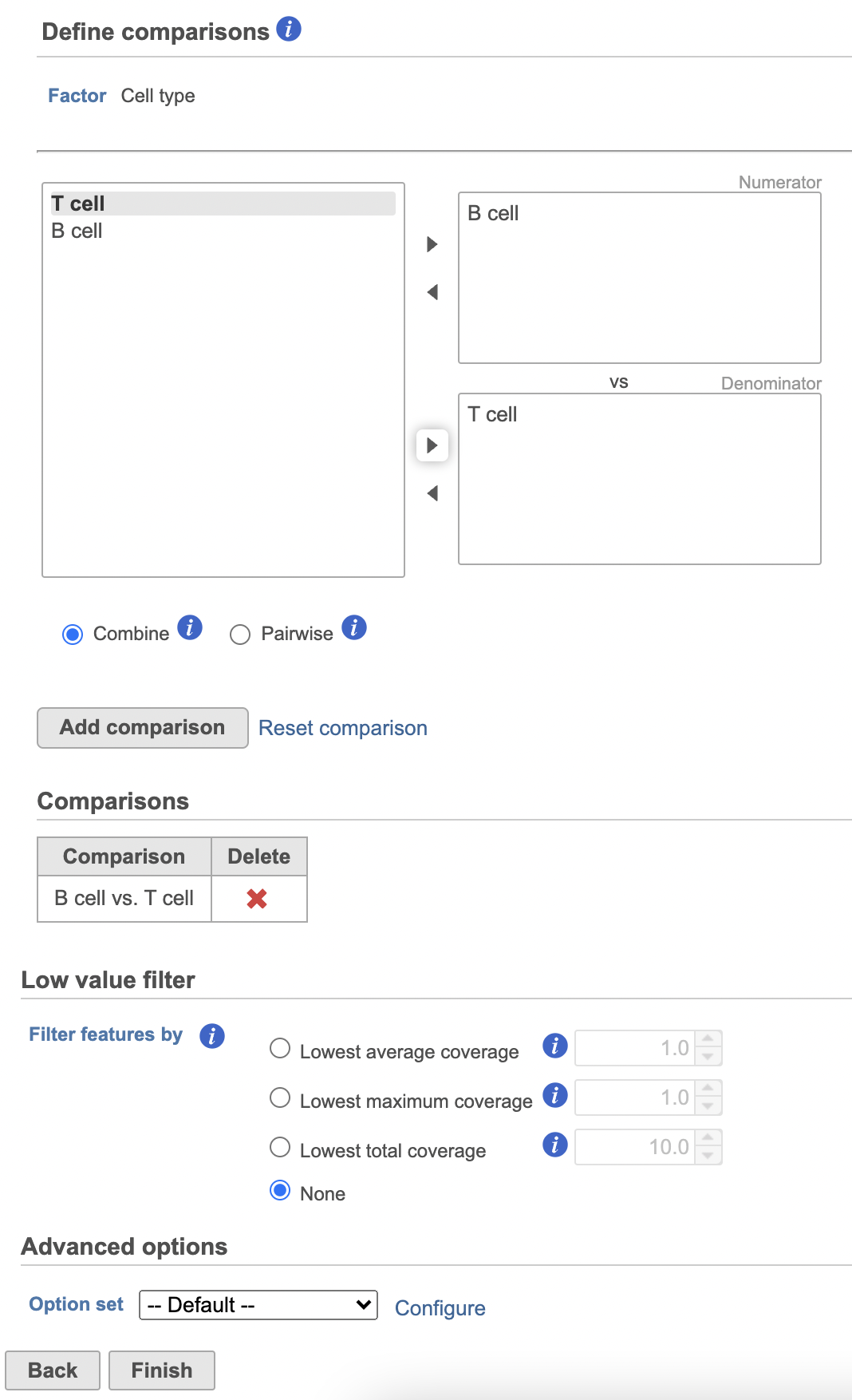 Image Removed Image Removed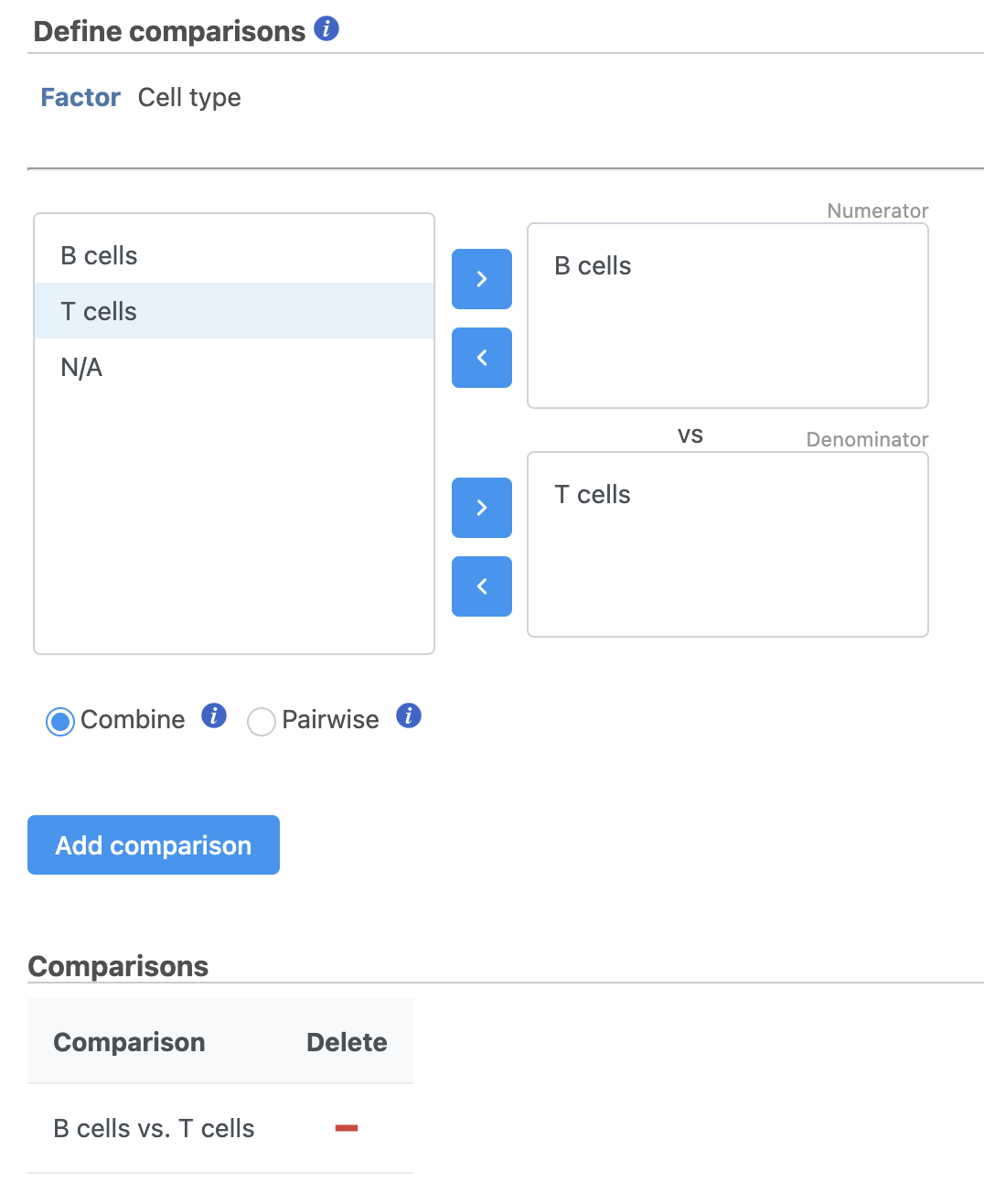 Image Added Image Added
|
Hurdle model produces a Feature list task node. The results table and options are the same as the GSA task report except the last two columns. The percentage of cells where the feature is detected (value is above the background threshold) in different groups (Pct(group1), Pct(group2)) are calculated and included in the Hurdle model report.
...
| Numbered figure captions |
|---|
| SubtitleText | Generate filtered node for differential analysis results in Flow. |
|---|
| AnchorName | Generate filtered node |
|---|
|
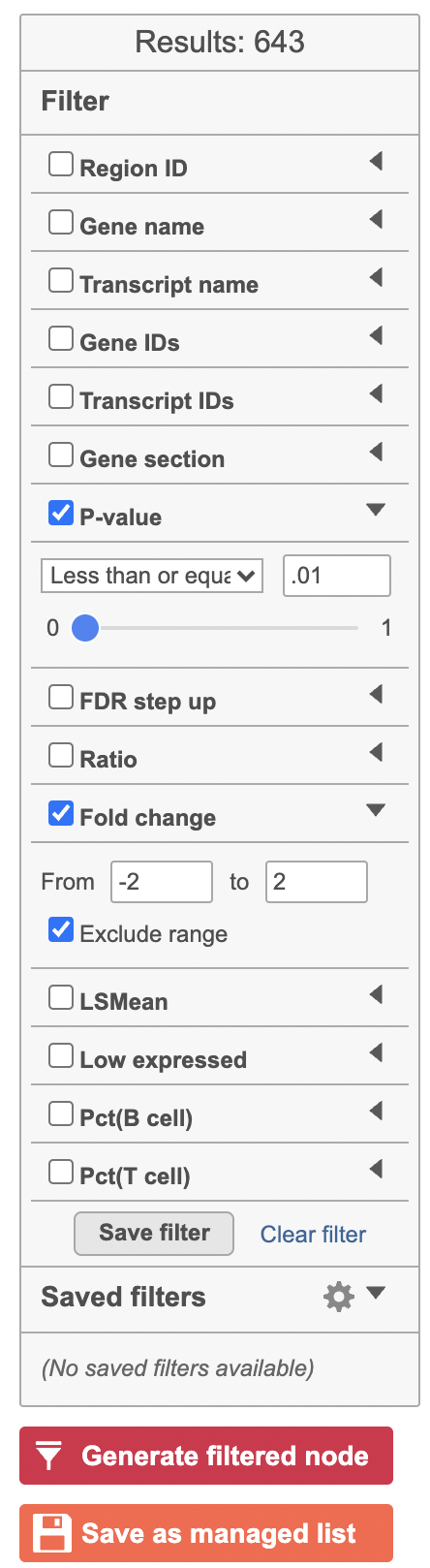 Image Removed Image Removed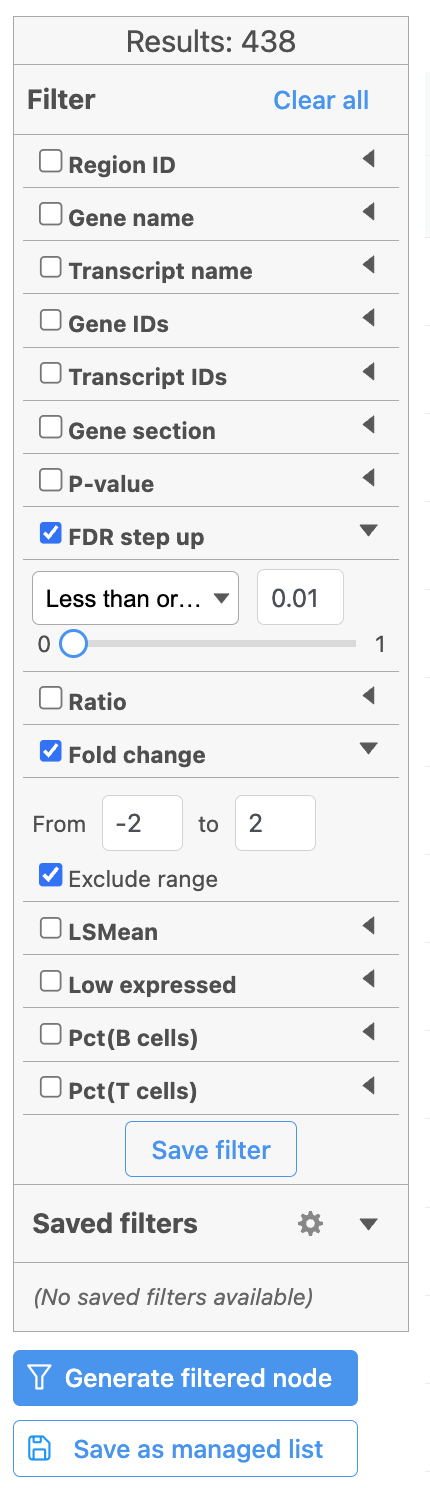 Image Added Image Added |
Once we have filtered a list of differentially expressed genes, we can visualize these genes by generating a heatmap, or perform the Gene set enrichment analysis and motif detection.
...
| Numbered figure captions |
|---|
| SubtitleText | Described pipeline shown in the Analyses tab |
|---|
| AnchorName | Pipeline as described |
|---|
|
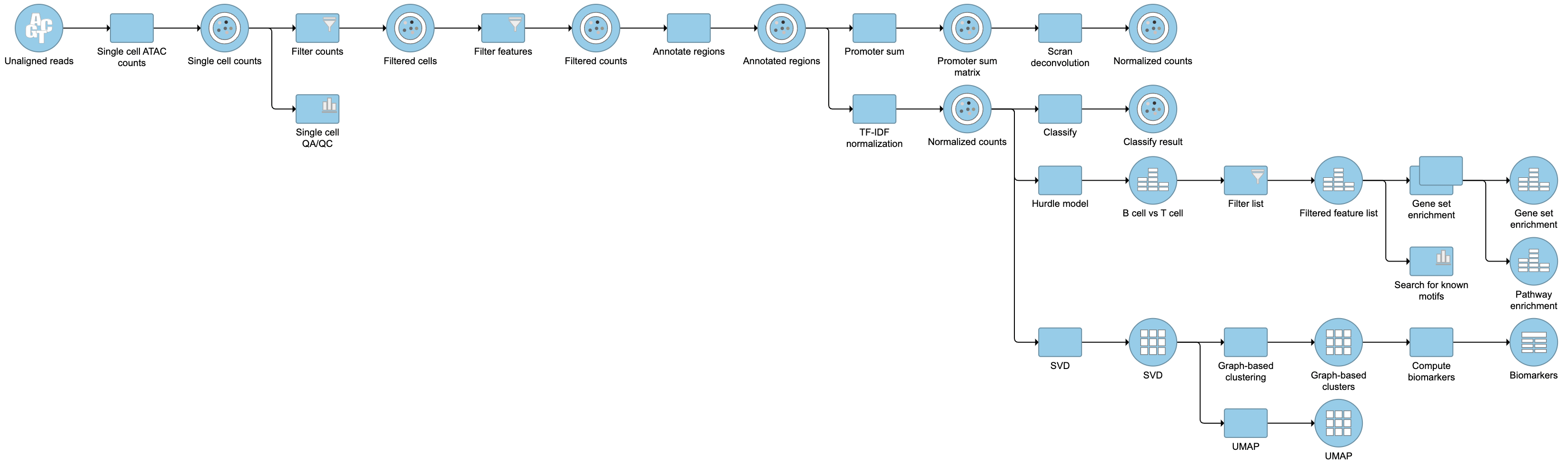 Image Modified Image Modified
|
For information about automating steps in this analysis workflow, please see our documentation page on Making a Pipeline.
...










































