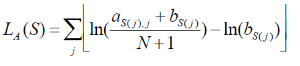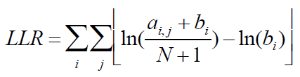Motif detection identifies enriched sequence motifs in peak regions generated by ChIP-Seq and ATAC-Seq data. Partek Flow includes Search for known motifs, which allows users to search for known motifs from a user-specified set or a database, and Detect de novo motifs, which can identify novel motifs. These tasks can be invoked on data nodes with genomic regions as features (not genes or transcripts).
Search for known motifs
Given a set of genomic regions, Search for known motifs can search for enrichment based on a sequence provided by user or using a sequence database like JASPAR.
- Click on a Peaks data node
- Click Motif detection in the toolbox
- Click Search for known motifs
The configuration dialog offers two search methods, By sequence and By database.
By sequence
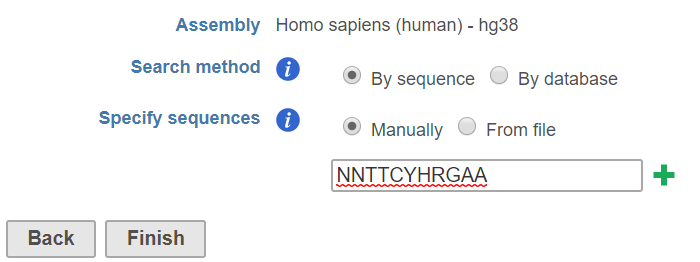
By sequence has two options (Figure 1):
Manually: the user can manually specify the sequences. Multiple sequences can be added by clicking the button
From file: use can specify a text file (.txt) that contains a list of sequences, one row per sequence.
The By sequence option uses a string search tool to return all positions in the set of genomic regions that match the given string(s). The string match is case insensitive, meaning if you search for ATCG, you may get atcG as a match. Nucleotides that are lower case have been "repeat masked", meaning they are located in a repetitive region of the genome. Your search string may contain any of the characters from the IUPAC nucleotide code . For example, if you search for WAAA, you may get back AAAA or TAAA (or any variation of upper and lower cases), because W represents A or T.
By database
The By database option uses an alignment matrix to match sequences against a motif database. We distribute the JASPAR database, but you can add any custom or public motif database.
Matching is determined using an alignment matrix. Alignment matrices are often used in literature to model transcription factor binding sites, alignment matrices are matrices of nucleotide counts per position [1]. Each instance of the motif is aligned to each other and the number of nucleotides at each position is counted and summarized in an alignment matrix. All positions from the set of genomic regions are scored against the alignment matrix. The score represents how likely the position is an instance of the motif. A quality cutoff is used to determine which sequences in the regions are instances of the motif. The scoring scheme and quality cutoff are similar to [2] and it briefly described below:
Let M be a motif of length L consisting of N motif instances.Let A be a 4XL alignment matrix such that ai,j is the count of letter i at position j. Let Bi be the background frequency of letter i (calculated as the number of nucleotides i in the regions divided by the total oligonucleotides in the regions). Let S be a sequence of length L. The score of S given the alignment matrix is
Equation 1
Let h be the maximum of LA. The quality score of a sequence is calculated as QA(S) =LA(S)/h. A quality score of 1 corresponds to a sequence with the most likely base at each position of the alignment matrix. The user will specify a threshold QA. All sequences that have a score TA>QA *h will be reported.
Task report
The Search for known motifs task report contains summary and detail tabs.
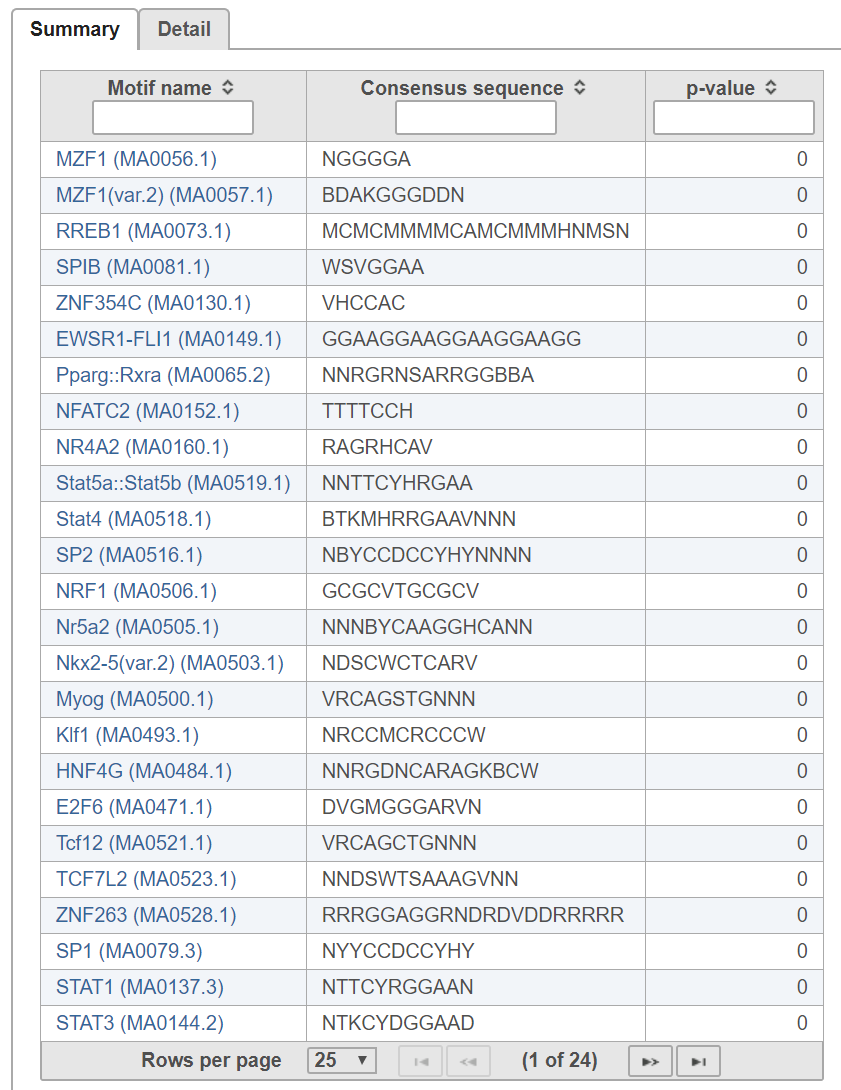
In the summary tab (Figure 2), each row is a motif in the search database specified. Clicking on the motif name opens the JASPAR database page with detailed information about the motif.
The p-value indicates whether instances of the motif are enriched in the input regions.
The p-value is calculated as follows:
The probability PExpected of a sequence having a score above TA is calculated under the assumption that the base are i.i.d. according to the background distribution B. Let NTrials be the number of sequences compared to the alignment matrix. The expected number of occurrences of the motif in the regions is PExpected * NTrials. The p-value of observing NActual instances with a score above TA is calculated based on the binomial distribution, where NTrials is the number of trials and PExpected is the probability of success.
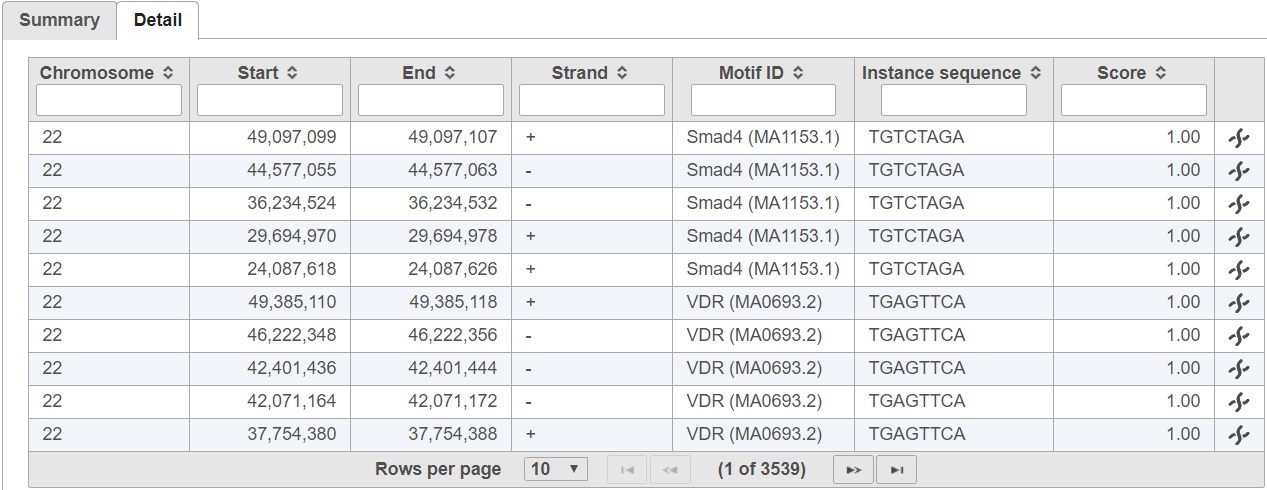
The detail tab lists motif sequence locations on rows and includes the quality score for each instance (Figure 3).
Detect de novo motifs
Detect de novo motifs can be used to identify novel motifs that are enriched in the input regions.
- Click on a Peaks data node
- Click Motif detection in the toolbox
- Click Detect de novo motifs
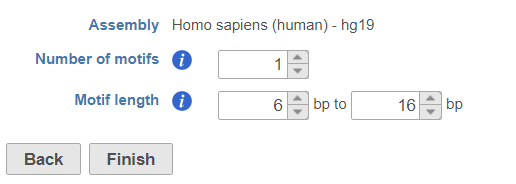
- Specify the number of motifs to report (Figure 4)
- Specify the range of motif length to search
Motif discovery uses Gibbs motif sampling. The implementation of the Gibbs motif sampling in Partek Flow is based on Neuwal, et all [3]. The Gibbs sampling method is a stochastic procedure that attempts to find the subset of sequences within the regions that maximizes the log likelihood ratio (LLR)
Equation 2
This is done by repeating the following two steps until convergence:
A. Given the alignment matrix from step B, search for location in the set of regions that score highly compared to the alignment matrix using Equation 1
B. Create a new alignment matrix from the set of high scoring positions from step A and return to step A.
The Gibbs sampler is run on a range of the motif sizes specified by the user. The motif with the greatest average LLR (LLR /length) is returned. To find N motifs in a set of regions, the Gibbs sampling method is run N times. The motif instances found from the previous run of the Gibbs sampler are removed before performing the next run.
Task report
The task report includes summary and detail tabs.
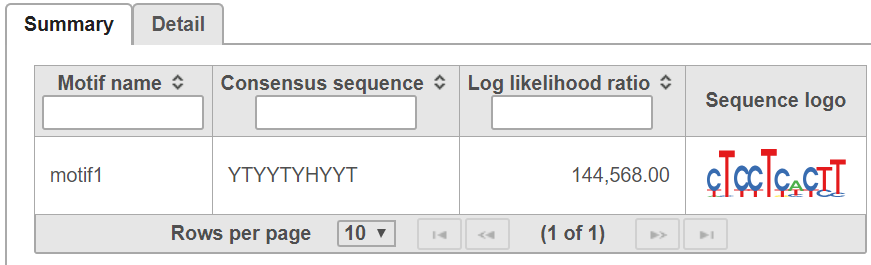
The summary tab gives the consensus sequence and sequence logo for each detected motif (Figure 5).
The detail tab is similar to the detail tab in Search for known motifs and lists the location of each motif sequence (Figure 6).
References
- Hertz, GZ., & Stormo, G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 1999, 15, 563-577
- Schug, J., & Overton, C.G. TESS, Transcription Element Search Software on WWW. Technical Report CBIL-TR-1997-1001-v0.0, of the Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania, 1997
- Neuwald, A.F., Liu, J.S., & Lawrence, C.E. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Science 1995, 4: 1618-1632.