Page History
Table of Contents maxLevel 2 minLevel 2 exclude Additional Assistance
| maxLevel | 2 |
|---|---|
| minLevel | 2 |
| exclude | Additional Assistance |
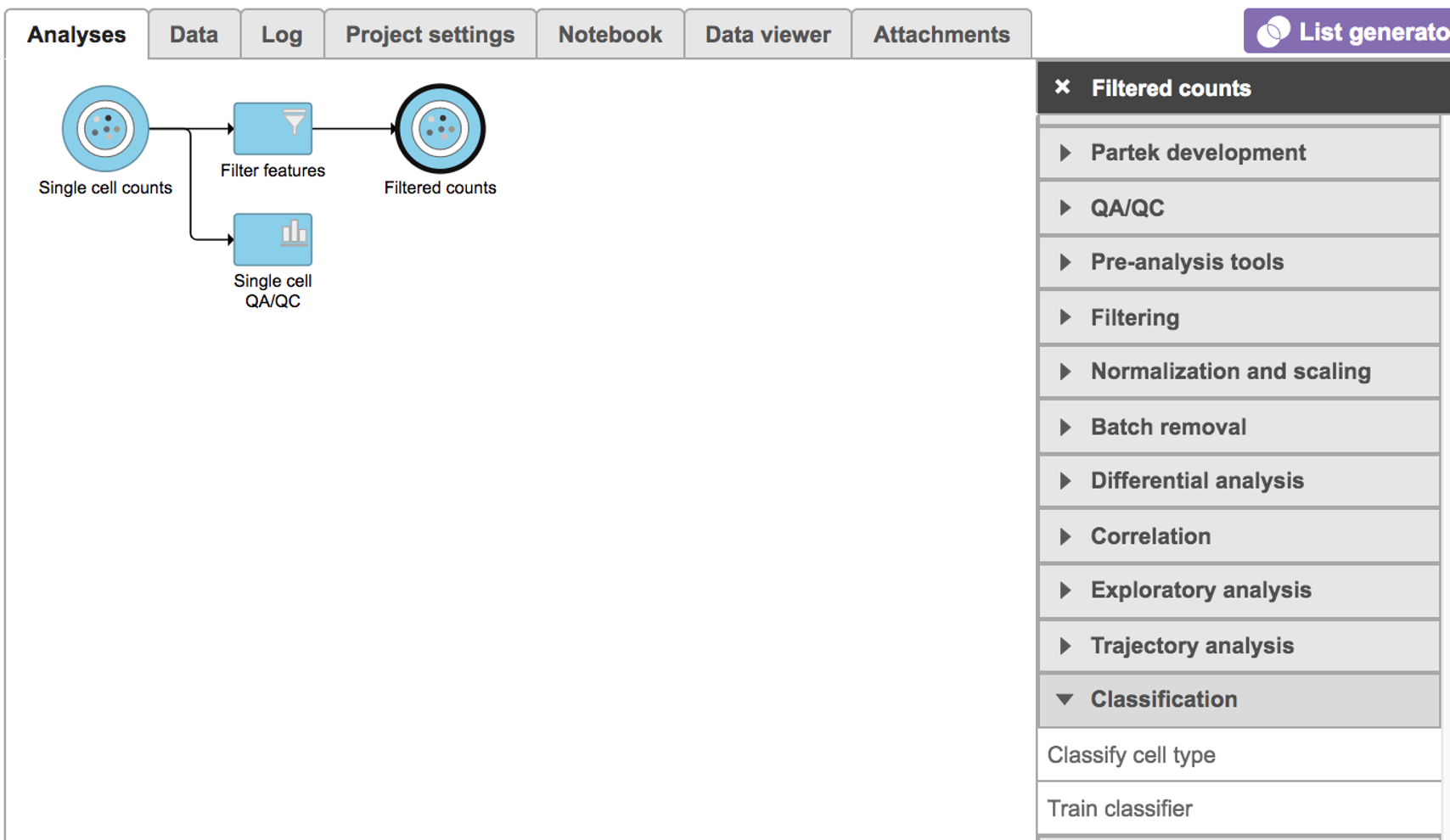
Classification in Partek Flow can be performed manually or with automatic cell classification.
Manual classification in Partek Flow
For more help related to manual classification please see Classifying cells or watch this related help video: Manual classification. Partek flow provides hosted lists from the literature for many cell types which can be added using List management.
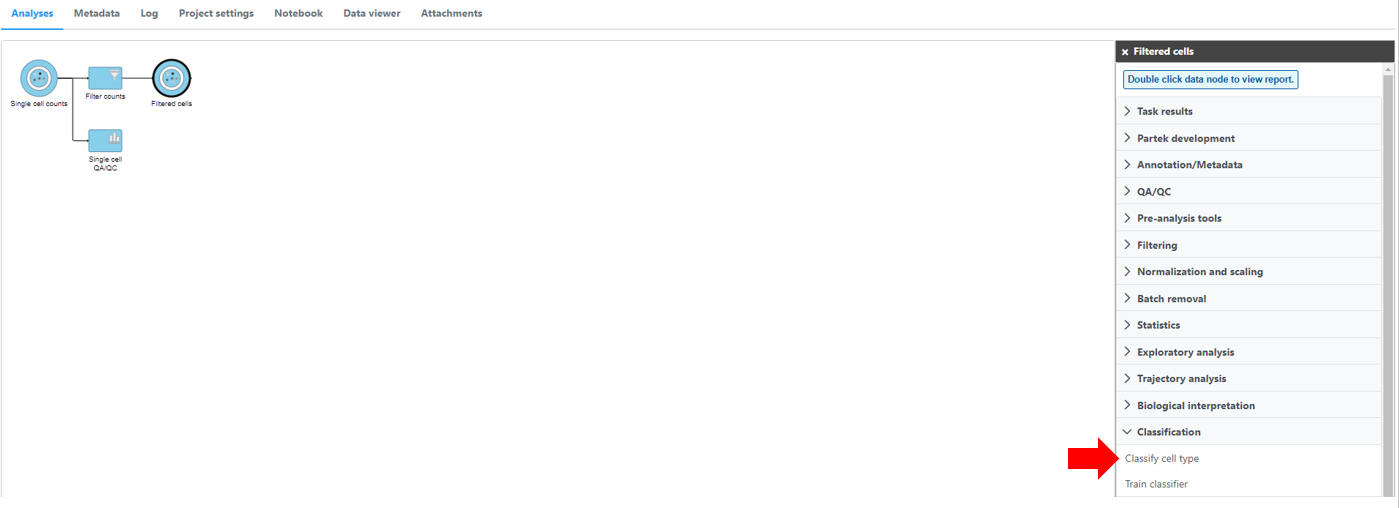
Automatic classification in Partek Flow
The Garnett1 automated cell type classification algorithm has been wrapped into Partek Flow as the Classification task. As with the original Garnett tool, Classification in Partek Flow works on single-cell data, along with a cell type definition (marker) file, and trains a regression-based classifier. Once a classifier is obtained and published, it can be applied to classify future datasets from similar tissues. To improve the user experience, both the marker file (.txt) and the classifier file (.rds) have been implemented as library files in Partek Flow.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
If using the Managed classifiers tool for the first time in Partek Flow, you will be asked to create a new classifier file (Figure 2a). Users may select either the Download Garnett classifier that matches the species and tissue type with their dataset from a Partek Flow maintained list or the Import Garnett classifier that is trained out of Partek Flow (Figure 2b). Next, push the Create button to create the classifier file. Once the correct classifier file has been created, select Finish to start running the task (Figure 2d).
...
Regardless of which type of classifier was applied, Partek Flow will output a new data node named Classify result (Figure 4). These outputs of cell type annotation are exactly the same as Garnett1. "cell_type" is the cell type assignments directly from Garnett model. While "cluster_ext_type" is the cell type that's determined by expanding cell type assignments to nearby cells using Louvain clustering. Downstream analysis tasks such as normalization and PCA can be performed on the Classify result data node.
...
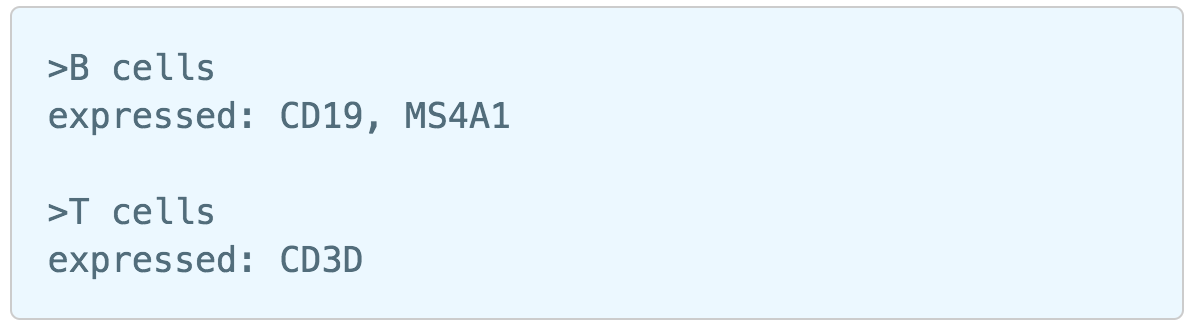
Similar to previously described steps, first time users will be asked to create the Marker file. Partek Flow does not currently host any marker files, however, users may add them as library files. Marker files should be a .txt file with the marker information in correct format. The same example in the Garnett tutorial of a simple valid Marker file is provided here (Figure 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Note:
1) Cell type name line starts with ">", followed by the name of the cell type, cell type name can NOT include hyphen (-).
2) Definition line starts with a keyword e.g. "expressed", "not expressed" eg. it is case sensitive. It followed by a ":" and space. Each gene name is followed by a comma and space, gene name is case sensitive. The line cannot be ended with comma or space.
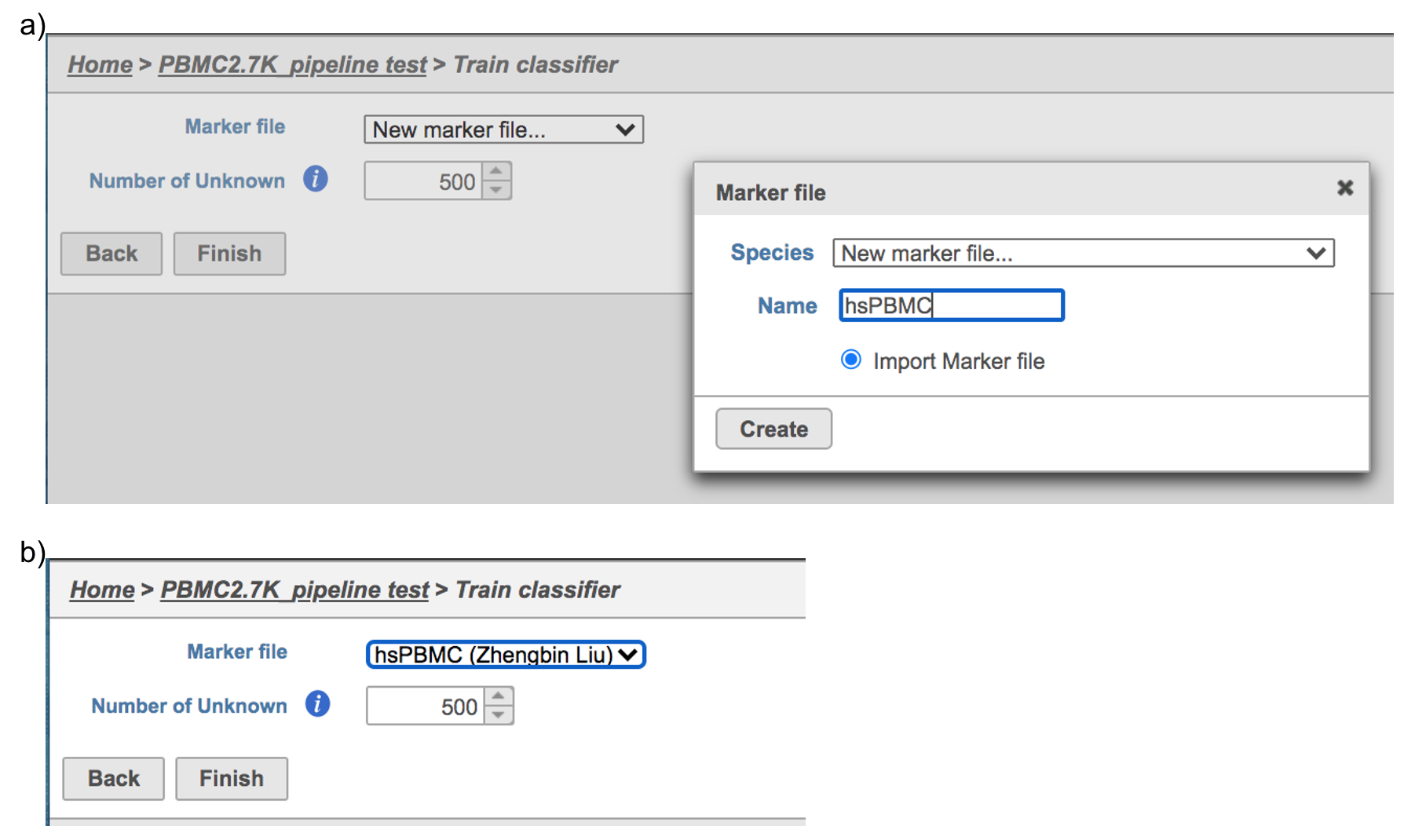
For more details on how to construct a Marker file, please refer to Garnett tutorial3. Next, click the Create button and Partek Flow will then save the file with the name that users provide for future use (Figure 6a).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
After the proper Marker file has been chosen, click the Finish button to start running the task as default (Figure 6b).
...
| Numbered figure captions | |||||||
|---|---|---|---|---|---|---|---|
| |||||||
|
Train classifier task report in Partek Flow
...