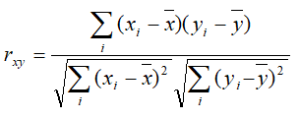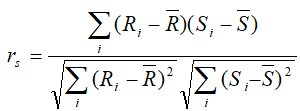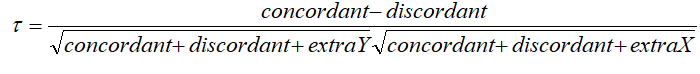Page History
Similarity matrix task is only available on bulk RNA-seq count matrix data node. It is used to compute the correlation of every sample/or feature vs every other sample/or feature. The result is a matrix with the same set of samples/or features on rows and columns, the value in the matrix is correlation coefficient --r.
Click on Similarity matrix task in Correlation section on the menu (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
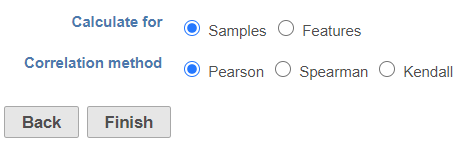
When the Sample correlation page opensthe dialog opens, you will be asked to select two samples for comparison select whether the calculation samples or features (Figure 2). The sample in the left box will be shown on the horizontal axis, while the sample in the right box will be shown on the vertical axis. Click on the sample names and then hit OK to proceed.are three correlation method options:
Pearson: linear correlation:
Spearman: rank correlation:
Kendal: rank correlation:
| Numbered figure captions | ||||||
|---|---|---|---|---|---|---|
|
| |||||
An example of the resulting scatterplot is in Figure 3. Each dot is a feature (gene/transcript) while the expression values in the two samples can be read off the coordinate axes, in the same units as present in the data node. For instance, if you normalized your RNA-seq data by transcripts per million (TPM), the coordinate axis will give you expression in TPMs. Pearson’s correlation coefficient and the slope of the regression line are in the upper left corner of the plot.
...