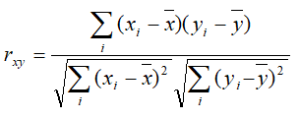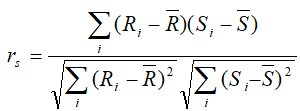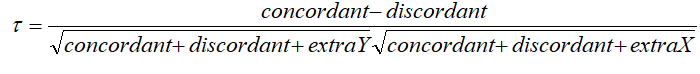Similarity matrix task is only available on bulk RNA-seq count matrix data node. It is used to compute the correlation of every sample/or feature vs every other sample/or feature. The result is a matrix with the same set of samples/or features on rows and columns, the value in the matrix is correlation coefficient --r.
Click on Similarity matrix task in Correlation section on the menu (Figure 1)
When the dialog opens, you will be asked to select whether the calculation samples or features (Figure 2). The are three correlation method options:
Pearson: linear correlation:
Spearman: rank correlation:
Kendal: rank correlation:
Click Finish to run the task. The output report of this task can be displayed in heatmap and/or table in the data viewer.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
12 | rates |