Page History
...
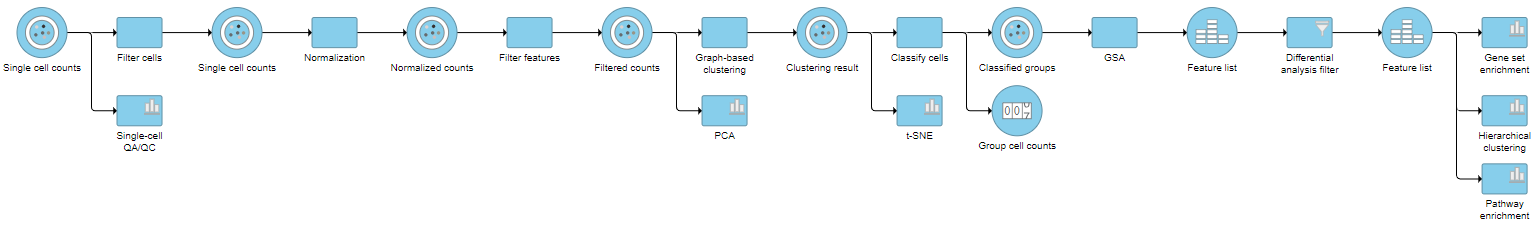
This tutorial includes only one sample, but the sames same steps will be followed when analyzing multiple samples. For notes on a few aspects specific to a multi-sample analysis, please see our Single Cell RNA-Seq Analysis (Multiple Samples) tutorial.
...
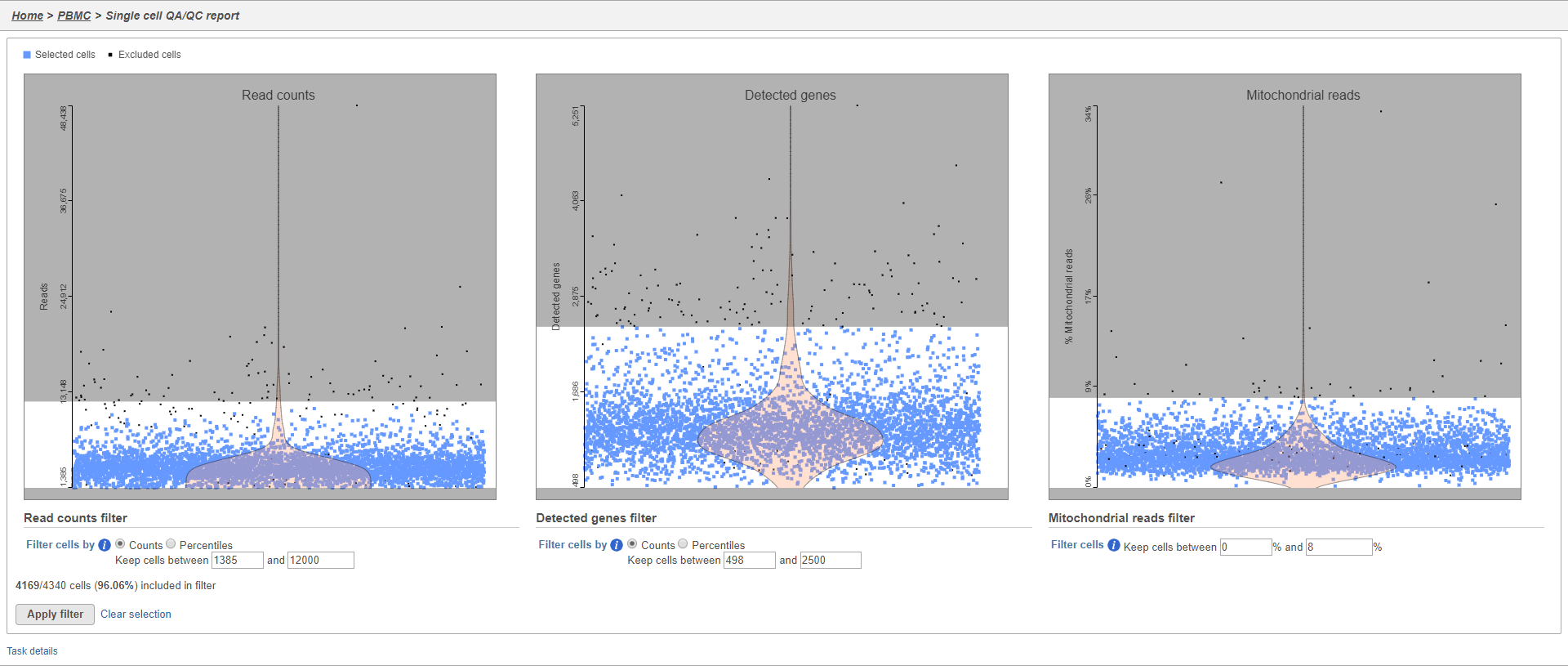
An important step in analyzing single cell RNA-Seq data is to filter out low quality cells. A few examples of low-quality cells are doublets, cells damaged during cell isolation, or cells with too few reads to be analyzed. You can do this in Partek Flow using the Single cell QA/QC task.
- Click on the Single cell data node
- Click on the QA/QC section of the task menu
- Click on Single cell QA/QC
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
There There are three plots: number of UMI read counts per cell, number of detected genes per cell, and the percentage of mitochondrial counts reads per cell.
Each point on the plots is a cell and the violins illustrate the distribution of values for the y-axis metric. Cells can be filtered either by clicking and dragging to select a region on one of the plots or by setting thresholds using the filters below the plots. Here, we will apply a filter for the number of read counts.
The plot will be shaded to reflect the filter. Cells that are excluded will be shown as black dots on both plots.
The UMI read counts per cell and number of detected genes per cell are typically used to filter out potential doublets - if a cell as an unusually high number of total UMIs counts or detected genes, it may be a doublet. The mitochondrial counts reads percentage can be used to identify cells damaged during cell isolation - if a cell has a high percentage of mitochondrial counts, it is likely damaged or dying and may need to be excluded.
...
Because different cells will have a different number of total UMIscounts, it is important to normalize the data prior to downstream analysis. For droplet-based single cell isolation and library preparation methods that use a 3' counting strategy, where only the 3' end of each transcript is captured and sequenced, we recommend the following normalization - 1. CPM (counts per million), 2. Add 1, 3. Log2. This accounts for differences in total UMI counts per cell and log transforms the data, which makes the data easier to visualize.
...
This produces a Filtered counts data node. This will be the starting point for the next stage of analysis - identifying cell types in the data using the interactive t-SNE plot.
Scaling
For some data sets, it may be necessary to remove technical artifacts or batch effects. To do this, you can use the Scaling task in the Normalization and Scaling section. To configure the scaling task, select the cell or sample attribute effects you would like to regress out of the data set. The scaling task is detailed in our Single Cell Scaling white paper. We will not perform scaling for this data set.
...
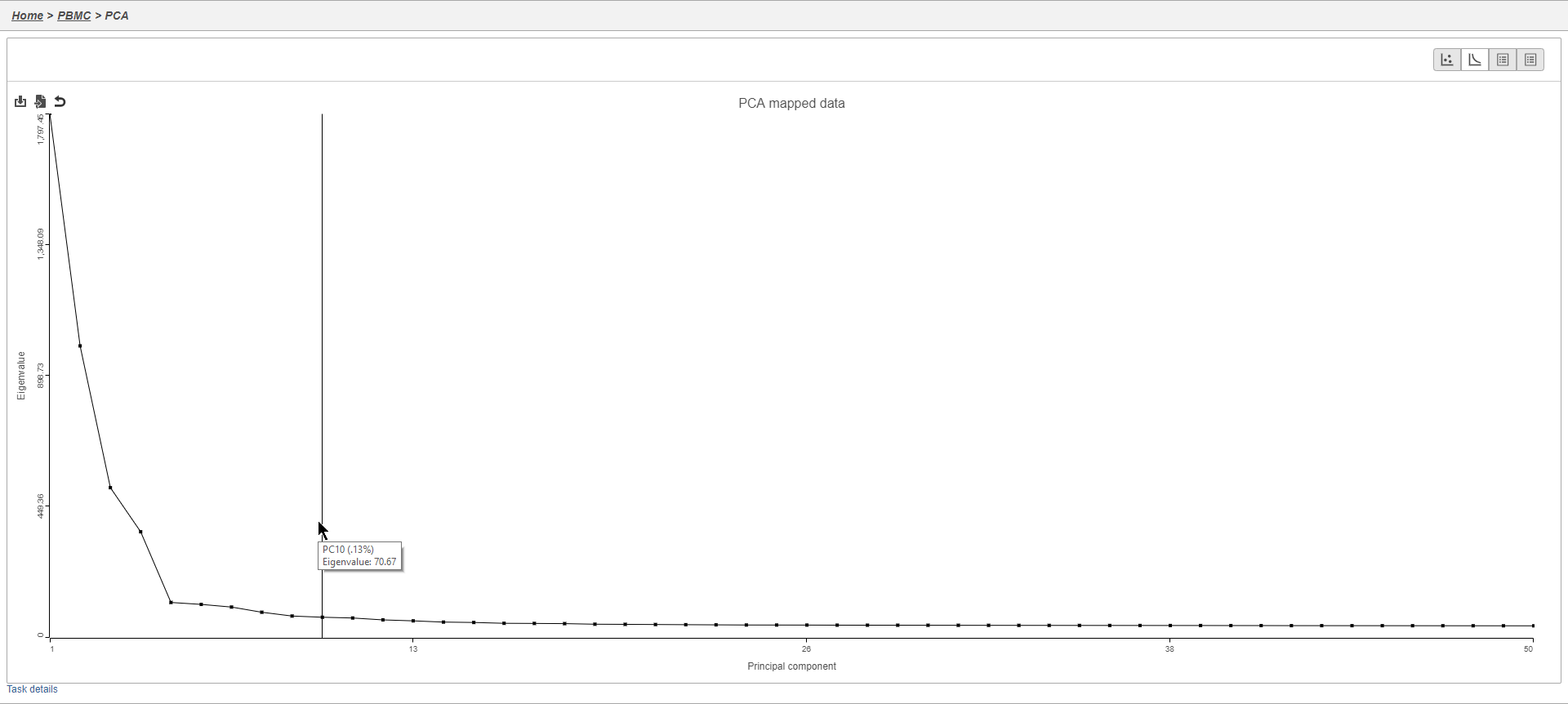
This will generate a Scree plot and PC component loadings table, which are useful for determining how many PCs to use for in downstream analysis tasks.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Viewing the genes highly correlated with each PC can be useful when choosing how many PCs to include.
...
In this case, we see a few known marker genes are highly correlated with PC1 (Figure 7).
...
Plot controls are located in the control panel on the left. You can adjust plot options, perform selection and filtering, and manage cell type classifications using the control panel. Below the scatter plot is the biomarker table that we saw in the the Graph-based clustering results. The table is interactive and clicking a gene in the table will color the plot by that gene (hold Ctrl on your keyboard and click to color by multiple genes in the table).
The t-SNE plot is in 3D by default. You can rotate the 3D plot by left-clicking and dragging your mouse. You can zoom in and out using your mouse wheel. You can pan by right-clicking and dragging your mouse. The 2D t-SNE is also calculated and you can switch between the 2D and 3D plots using the Plot style radio buttons in the control panel. In 3D, you can switch from points to 3D spheres and also add a fog effect to improve depth perception on the plot. To produce an optimal plot, you can also adjust size of the points using the Point size slider.
Coloring the t-SNE scatter plot
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
If you want to color by more than three genes at time, for example, by a list of genes that distinguish a particular cell type, like B cells, you can use the color by list option.
...
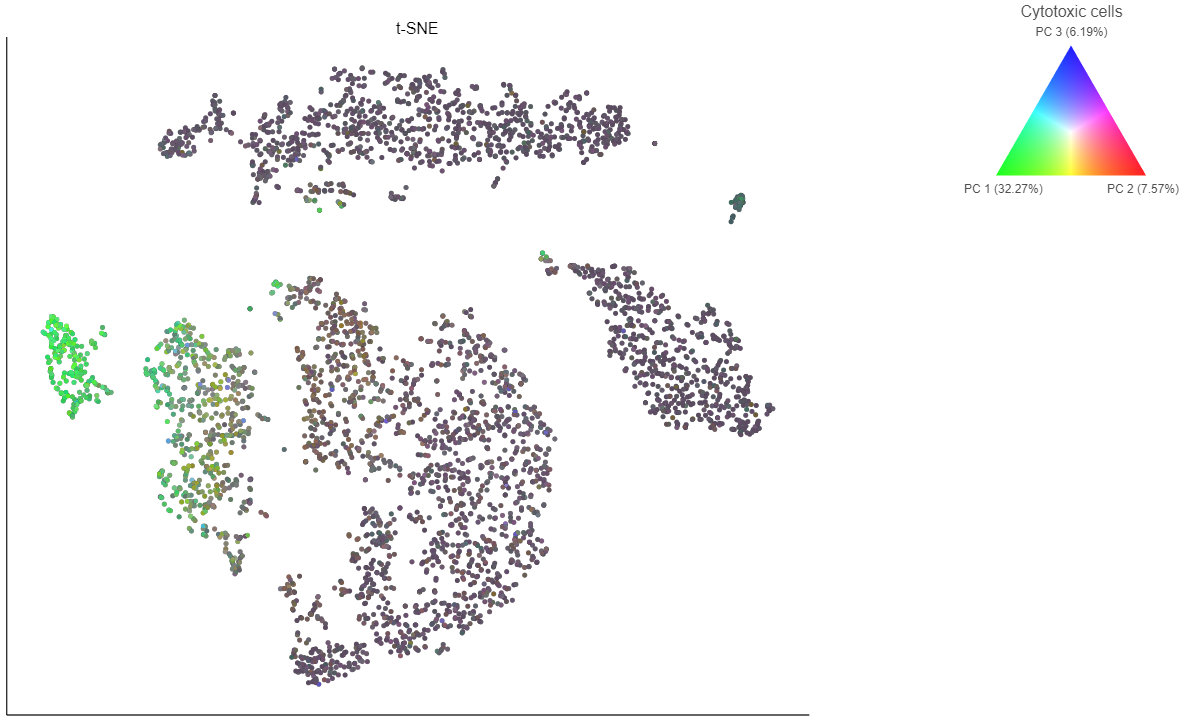
Coloring by a list calculates the first three principal components for the gene list and color the cells on the plot by their values along those three PCs with green for PC1, red for PC2, and blue for PC3 (Figure 16).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
In addition to coloring by gene expression and by gene lists, the points can be colored by any cell or sample attribute. Each of the Available attributes is are listed as an option options in the Color by drop-down menu.
...
- Click to activate Lasso mode
- Left-click and hold to draw a lasso around a cluster of cells
- Release and click the starting circle to close the lasso and select the enclosed cells (Figure 17)
You can also create a lasso with straight lines using Lasso mode by clicking, releasing, and clicking again to draw a shape.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
By default, selected cells are shown in bold while unselected cells are dimmed (Figure 18). This can be changed to gray unselected selected cells using the Selection/Filtering section of the control panel.
...
Classifying cells allows to you assign cells to groups that can be used in downstream analysis and visualizations. Commonly, this is used to describe cell types, such as B cells and T cells, but can be used to describe any group of cells that you want to consider together in your analysis, such as cycling cells or CD14 high expressing cells. Each cell can only belong to one class at a time so you cannot create overlapping classes.
To classify a cellscell, just select it then click Classify selection.
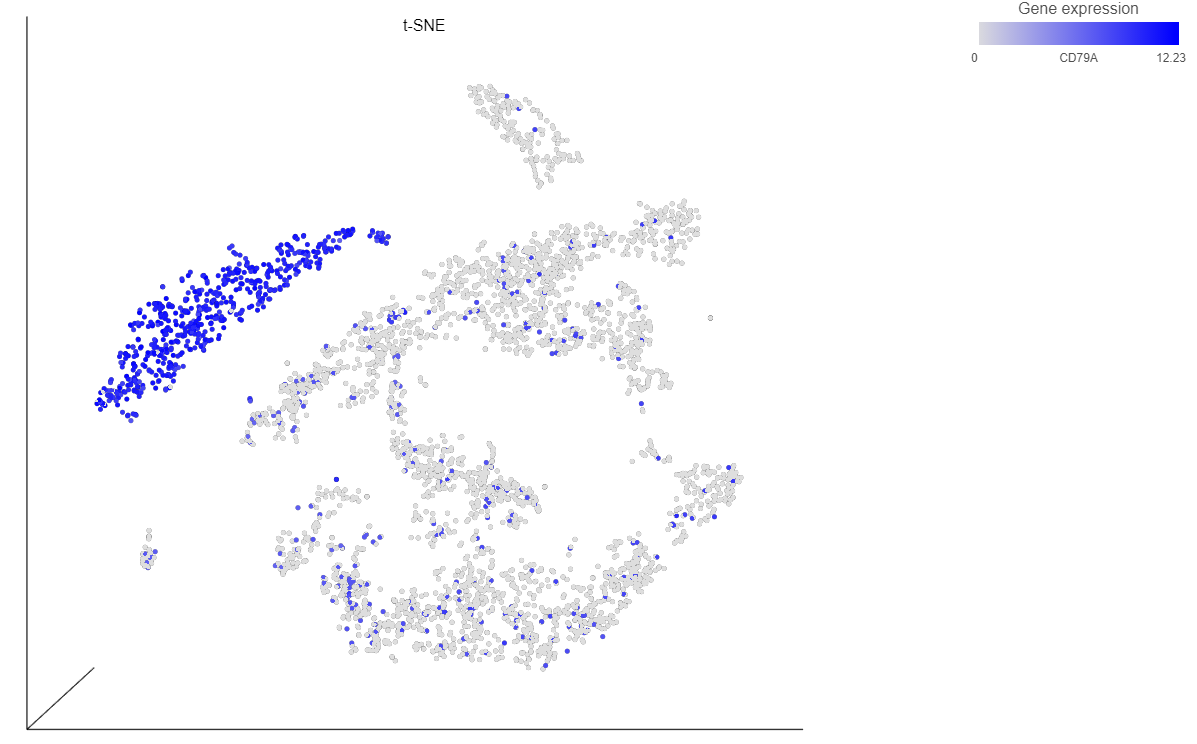
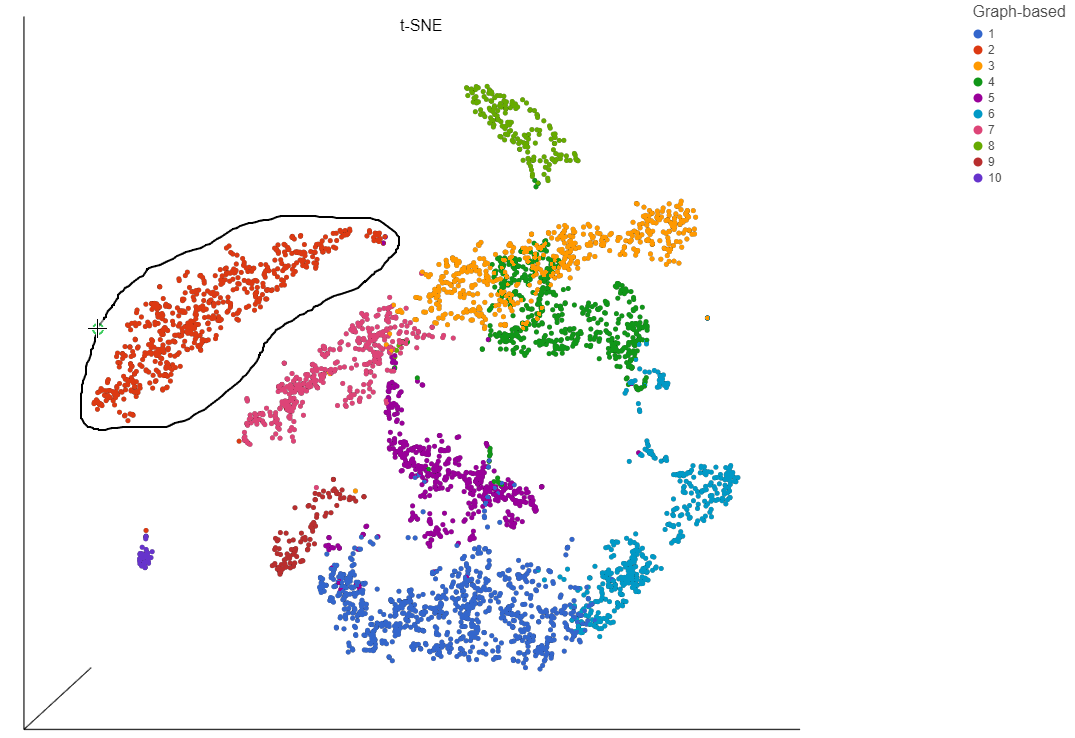
As an For example, we can classify a cluster of cells expressing high levels of CD79A as B - cells.
- Set Color by to Gene expression
- Type CD79A in the Gene expression search box and select it
- Click to activate Lasso mode
- Draw a lasso around the cluster of CD79A-expressing cells (Figure 24)
...
Because most of these cells express CD79A, a B cell marker, and because they cluster together on the t-SNE, suggesting they have similar overall gene expression, we believe that all these cells are B cells.
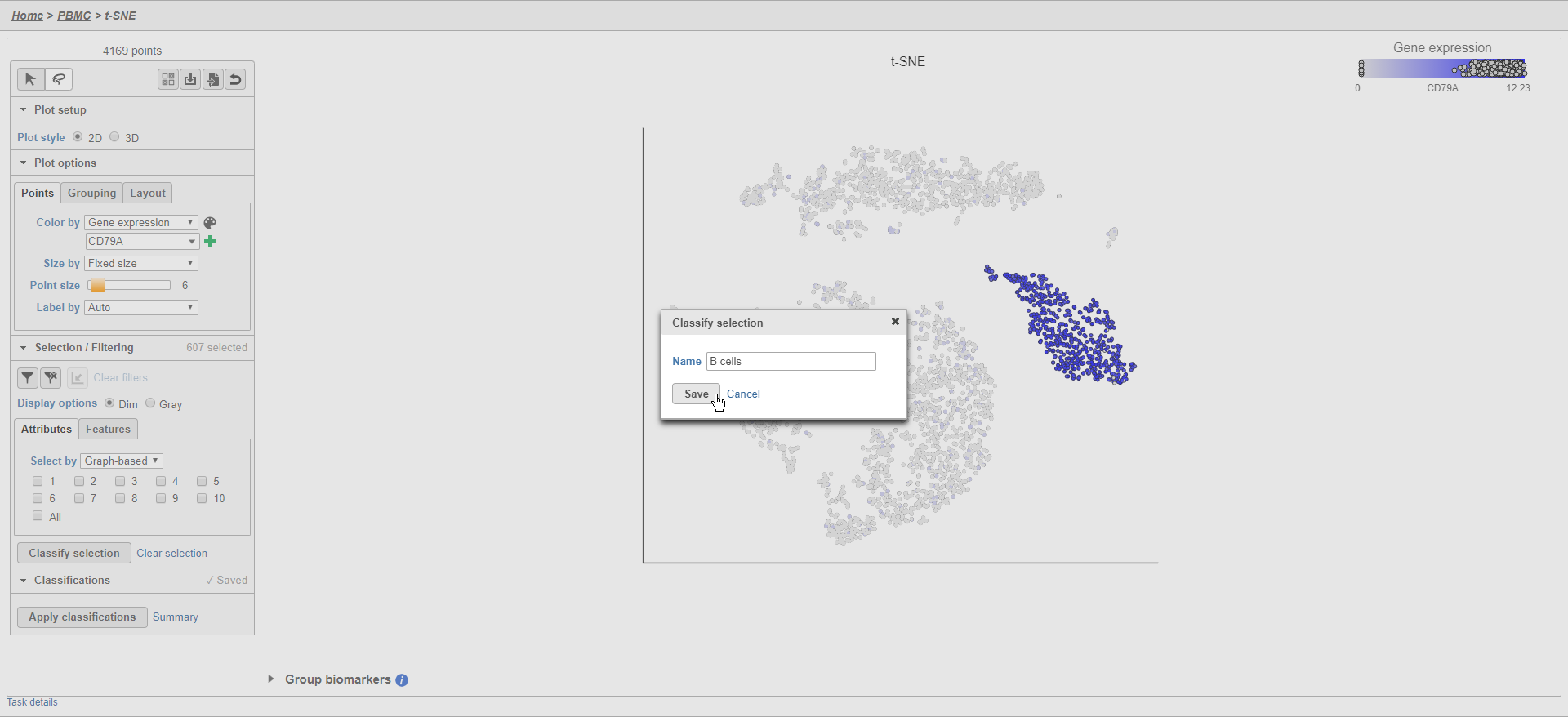
- Click Classify selection
- Type B cells for the Name
- Click Save (Figure 25)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The classification classification, B cells, is added to the Classifications section of the control panel and the number of cells in that class classification is listed next to the name (Figure 26).
...
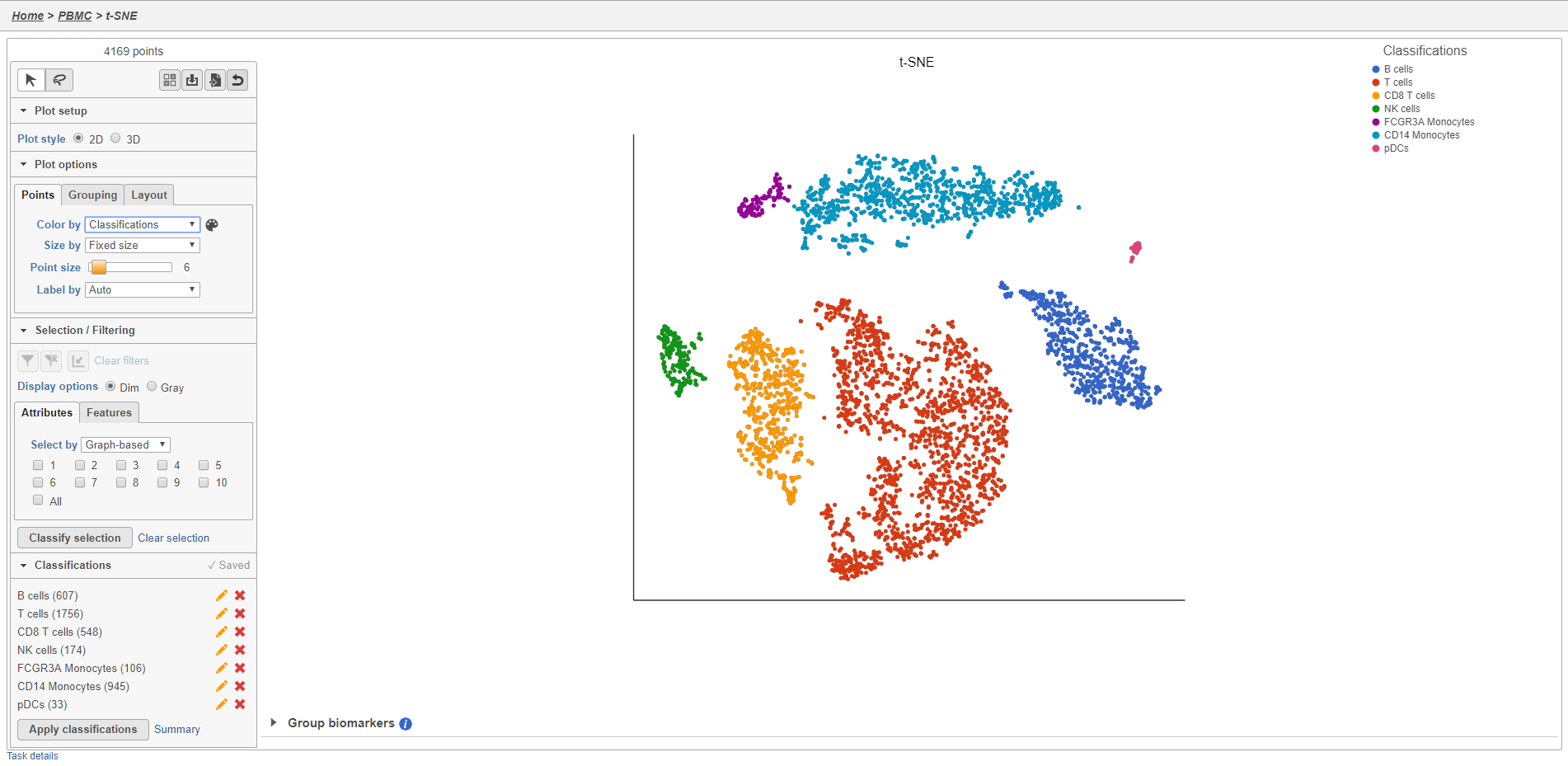
You can edit the name of a classification by selecting or delete it by selecting . The classification state of classifications you have made on the scatter plot is are saved as a working draft so if you close the plot and return to it, the saved classifications will still be there. However, classifications are not available for downstream tasks until you apply them.
Once you have added saved a classification, you can color the t-SNE plot by the new attribute Classification. Here, I classified a new few additional cell types using a combination of known marker genes and the clustering results and colored the plot by Classification (Figure 27).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
The number of cells for each sample is listed (Figure 31). This data node can be used as the starting point for differential cell count analysis. This is particularly useful in multi-sample experiments, such as immunotherapy studies, where the number of cells of a particular cell type if in two or more conditions is of interest.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
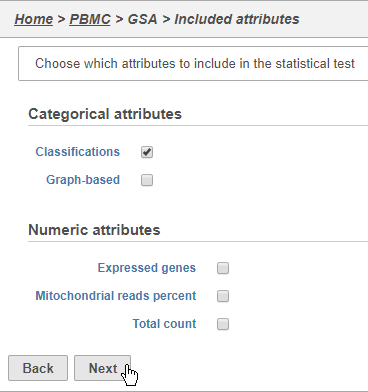
Here, we We will make a comparison between NK cells and all the other cell types to identify genes that distinguish NK cells. However, you could also use this tool to identify genes that differ between two cell types or genes that differ in the same cell type between experimental conditions.
...
The top panel is the numerator for fold-change calculations so the experimental or test group groups should be selected in the top panel.
...
The GSA task report lists genes in on rows and the results of the statistical test (p-value, fold change, etc.) on columns (Figure 34). For more information, please see our documentation page on the GSA task report.
...
The GSA report will close and a new task, the Differential analysis filter, will run and generate a filtered Feature list data node.
For more information about the GSA task, please see the Differential Gene Expression - GSA section of our user manual.
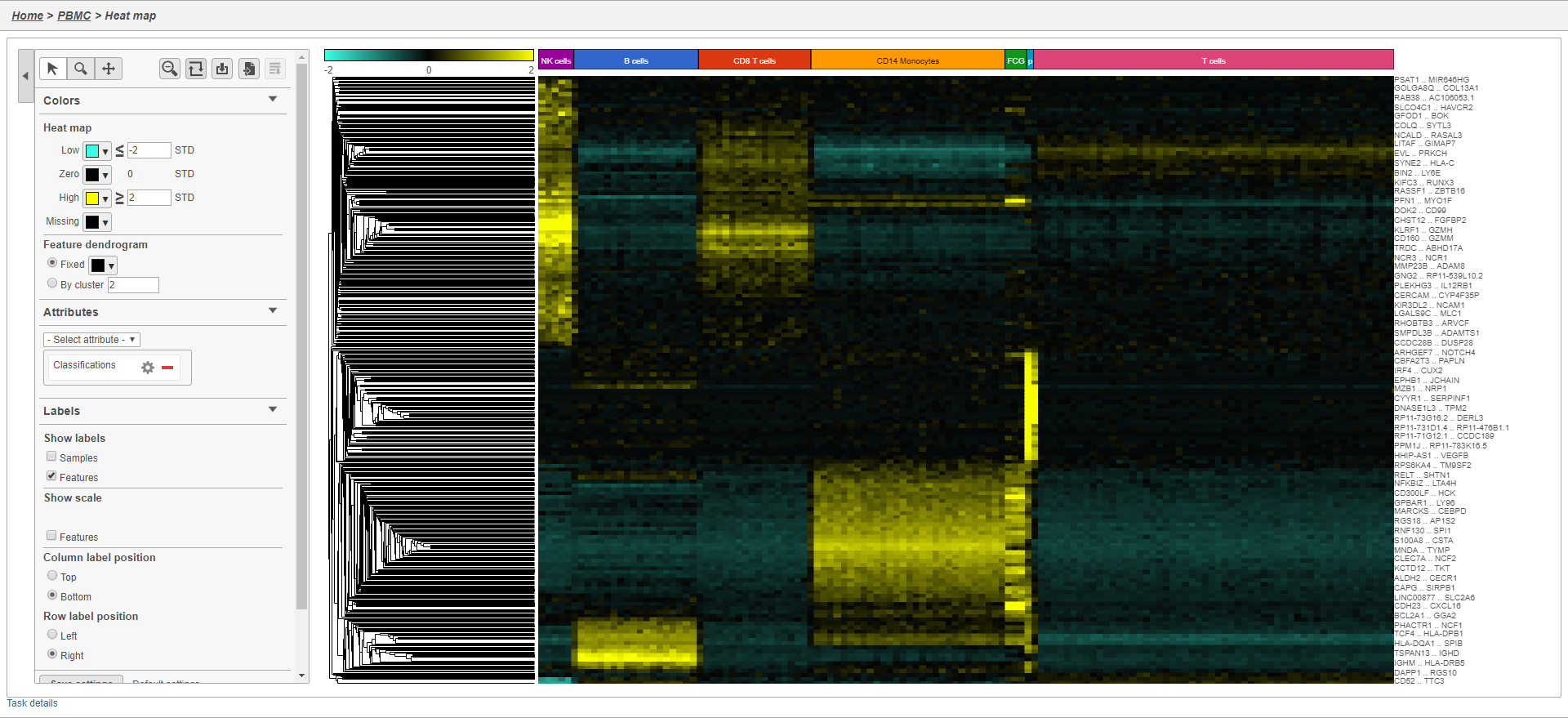
Generating a heat map
Once we have filtered to a list of significantly different genes, we can visualize these genes by generating a heat map.
...
The heat map now shows a teal to yellow gradient with a black midpoint (Figure 38).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
As with any visualization in Partek Flow, the image can be saved as a publication-quality image to your local machine by clicking or sent to a page in the project notebook by clicking . For more information about Hierarchical clustering, please see the Hierarchical Clustering section of the user manual.
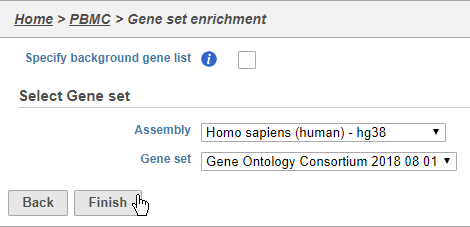
Performing enrichment analysis
...
- Choose Homo sapiens (human) - hg38 from the Assembly drop-down
- Choose Gene Ontology Consortium 2018 08 01 from the Gene set drop-down
- Click Finish (Figure 39)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
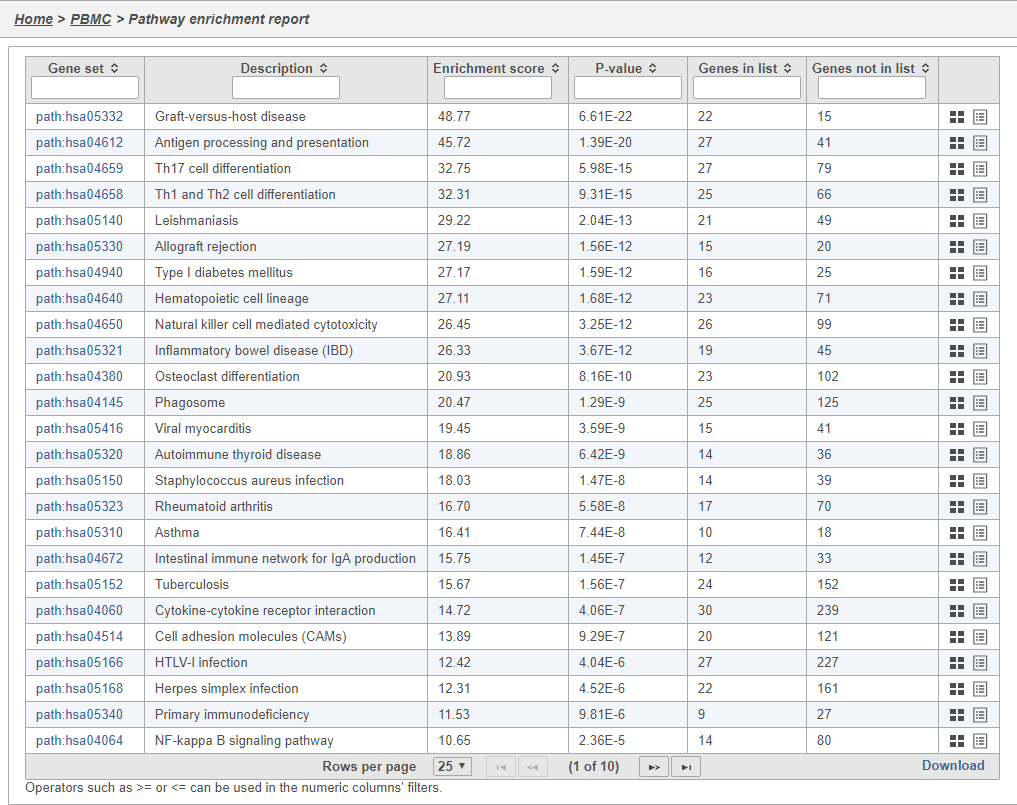
In Partek Flow, you can also check for enrichment of KEGG pathways using the Pathway enrichment task. The task is quite similar to the Gene set enrichment task, but uses KEGG pathways as the gene sets.
Clicking the KEGG pathway ID in the Pathway enrichment task report The task report is similar to the Gene set enrichment task report with enrichment scores, p-values, and the number of genes in and not in the list (Figure 41). opens a KEGG pathway map.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
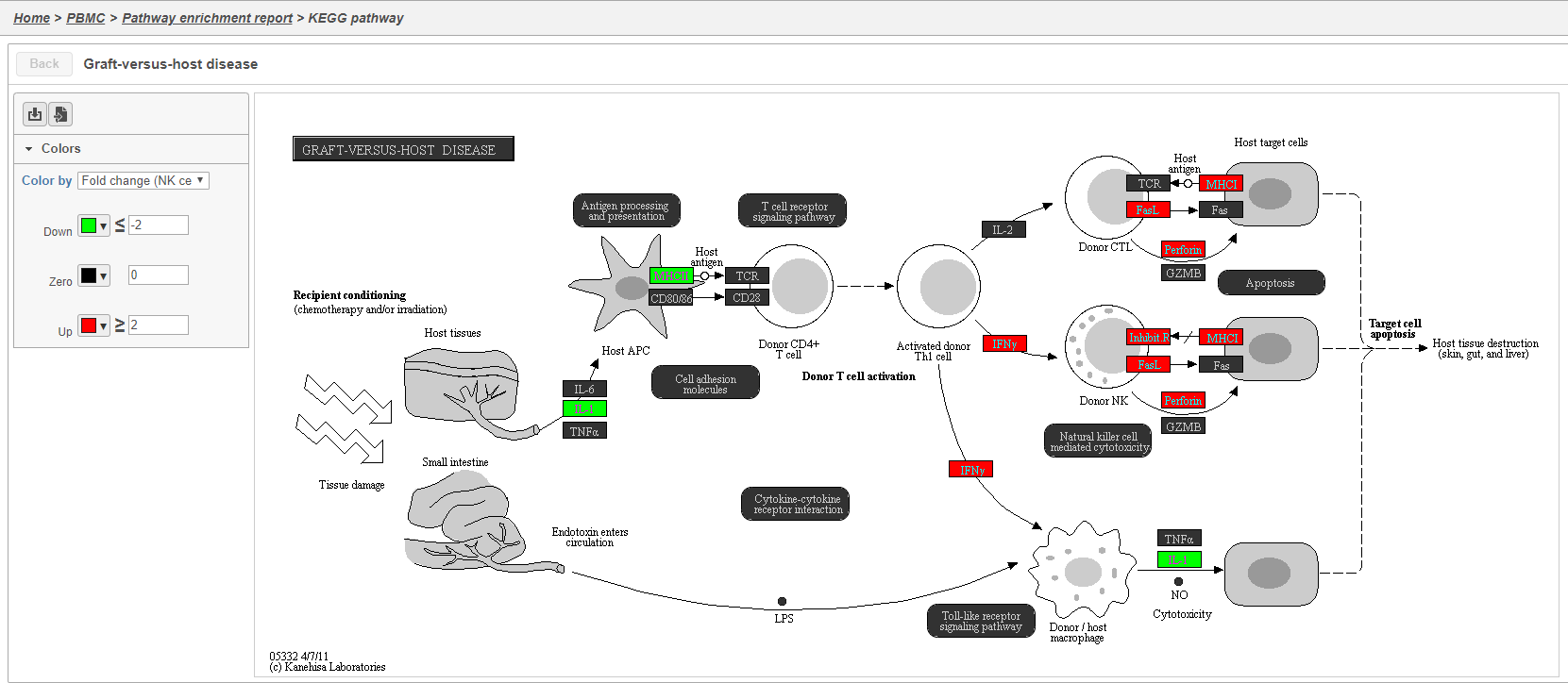
The Clicking the KEGG pathway ID in the Pathway enrichment task report opens a KEGG pathway map (Figure 42). The KEGG pathway maps have fold-change and p-value information from the input gene list overlaid on the map, adding a layer of additional information about whether the pathway was upregulated or downregulated in the comparison (Figure 42).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
For information about automating steps in this analysis workflow, please see our documentation page on Making a Pipeline.
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|