This tutorial presents an outline of the basic series of steps for analyzing a single cell RNA-Seq experiment in Partek Flow starting with the count matrix file.
This tutorial includes only one sample, but the same steps will be followed when analyzing multiple samples. For notes on a few aspects specific to a multi-sample analysis, please see our Single Cell RNA-Seq Analysis (Multiple Samples) tutorial.
If you are new to Partek Flow, please see Getting Started with Your Partek Flow Hosted Trial for information about data transfer and import and Creating and Analyzing a Project for information about the Partek Flow user interface.
Filtering cells
An important step in analyzing single cell RNA-Seq data is to filter out low quality cells. A few examples of low-quality cells are doublets, cells damaged during cell isolation, or cells with too few reads to be analyzed. You can do this in Partek Flow using the Single cell QA/QC task.
- Click on the Single cell data node
- Click on the QA/QC section of the task menu
- Click on Single cell QA/QC
A task node, Single cell QA/QC, is produced. Initially, the node will be semi-transparent to indicate that it has been queued, but not completed. A progress bar will appear on the Single cell QA/QC task node to indicate that the task is running (Figure 1).
- Click the Single cell QA/QC node once it finishes running
- Double-click the Task report in the task menu
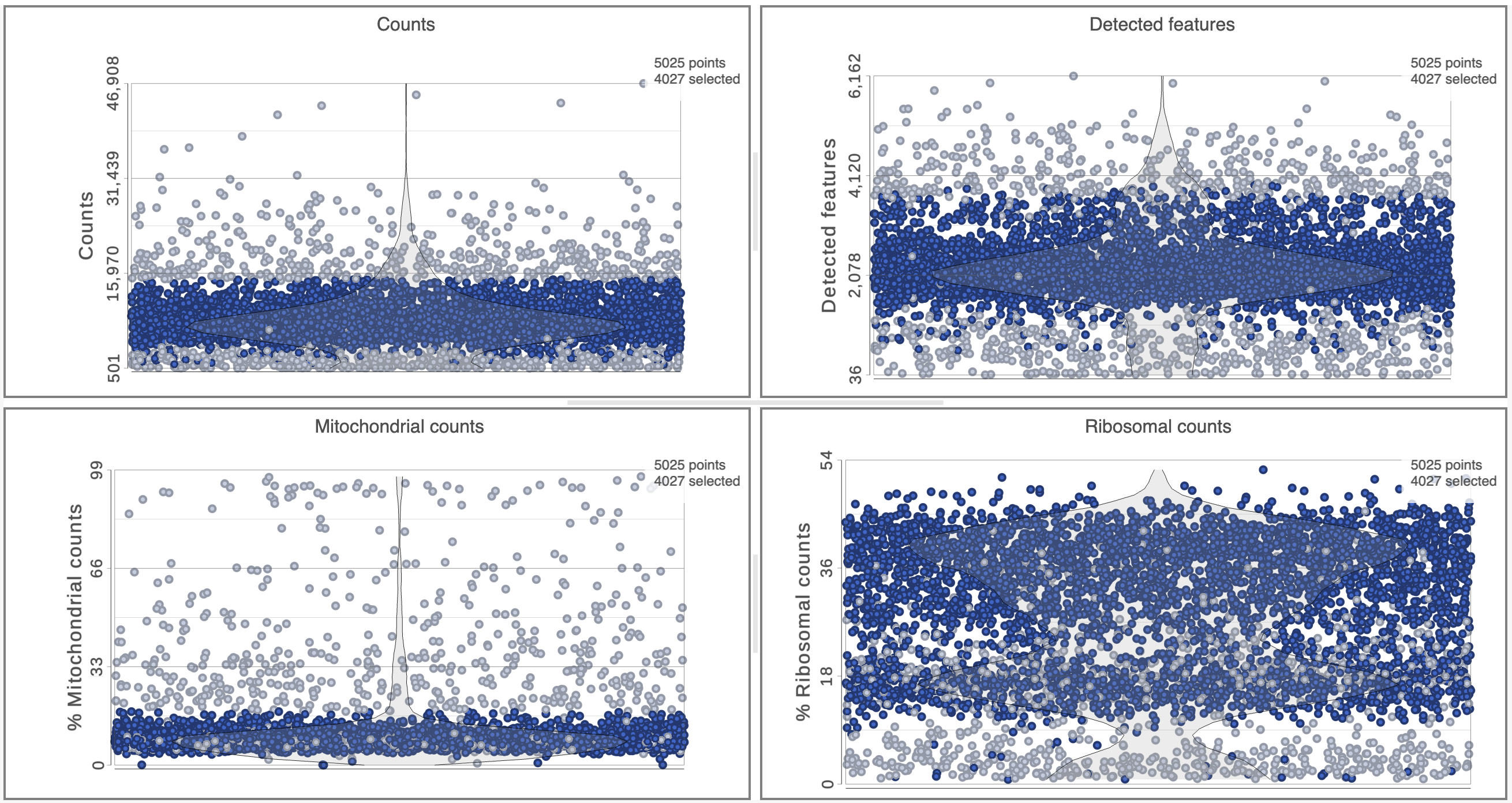
The Single cell QA/QC report includes interactive violin plots showing the value of every cell in the project on several quality measures (Figure 2).
There can be four plots: number of read counts per cell, number of detected genes per cell, the percentage of mitochondrial reads per cell, and the percentage of ribosomal counts.
Each point on the plots is a cell and the violins illustrate the distribution of values for the y-axis metric. Cells can be filtered either with the plot controls by using the selection tools on the right of the plot (rectangle mode , ellipse mode , or lasso mode ) and selecting a region on one of the plots or by setting thresholds using the Select & Filter tool. Here, we will apply a filter for the number of read counts.
The plots will be shaded to reflect the selection. Cells that are excluded will be shown as dim dots on all plots.
The read counts per cell and number of detected genes per cell are typically used to filter out potential doublets - if a cell as an unusually high number of total counts or detected genes, it may be a doublet. The mitochondrial reads percentage can be used to identify cells damaged during cell isolation - if a cell has a high percentage of mitochondrial counts, it is likely damaged or dying and may need to be excluded.
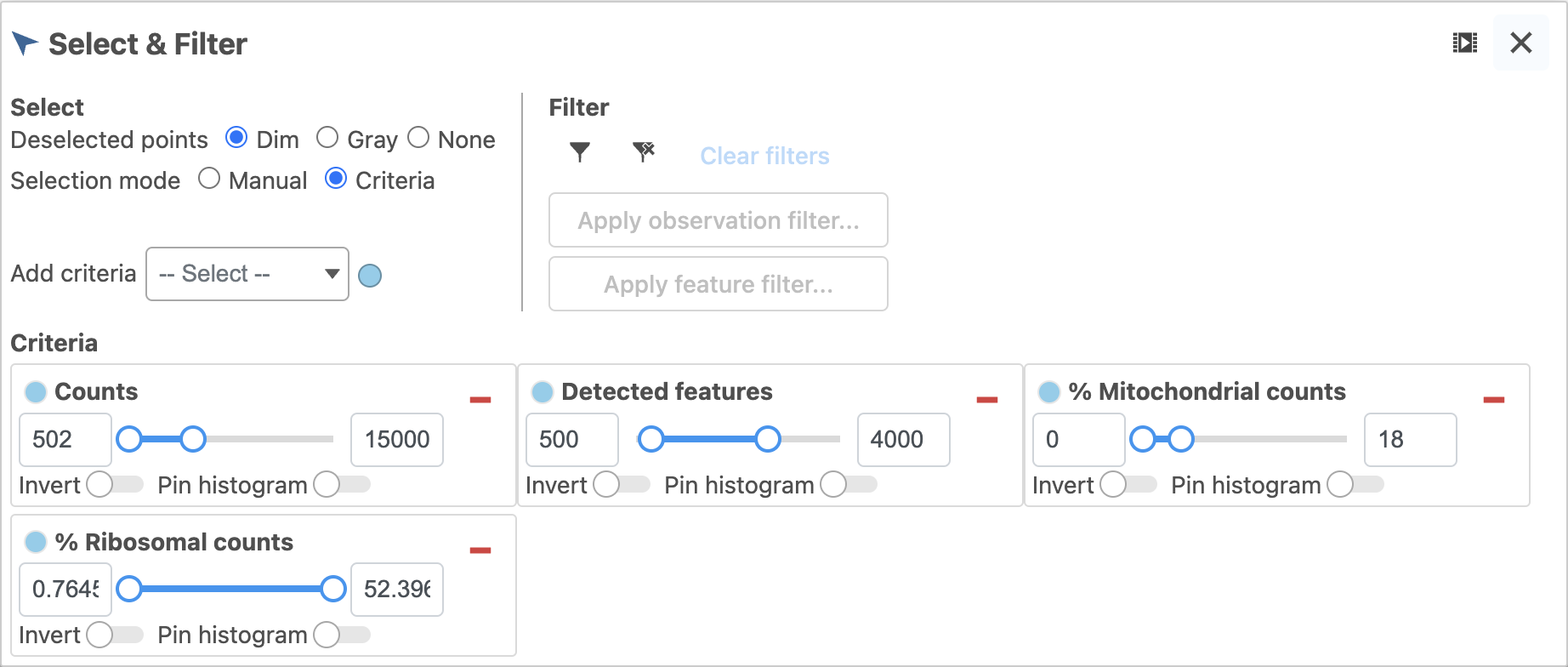
- Open the Select & Filter icon in the left panel. The histograms can be pinned while fine tuning the selections (Figure 2). Set the filters to represent the majority of the population (violin width)
- Click the filter icon and Apply observation filter then select the Single cell counts data node to run the Filter cells task on the first Single cell counts data node, it generates a Filtered counts task node that generates a Filtered cells results node
- Use Save as to give this Data Viewer session a new name (e.g. QA/QC filter) so you can return to this filter at any time and see the exact criteria that has been selected and filtered.
Filter features
A common task in bulk and single-cell RNA-Seq analysis is to filter the data to include only informative genes (features). Because there is no gold standard for what makes a gene informative or not and ideal gene filtering criteria depends on your experimental design and research question, Partek Flow has a wide variety of flexible filtering options. The Filter features step can also be performed before normalization or after normalization.
- Click the data node containing count matrix
- Click Filtering in the task menu
- Click Filter features
There are four categories of filter available - noise reduction, statistics based, feature metadata, and feature list.
The noise reduction filter allows you to exclude genes considered background noise based on a variety of criteria. The statistics based filter is useful for focusing on a certain number or percentile of genes based on a variety of metrics, such as variance. The feature list filter allows you to filter your data set to include or exclude particular genes.
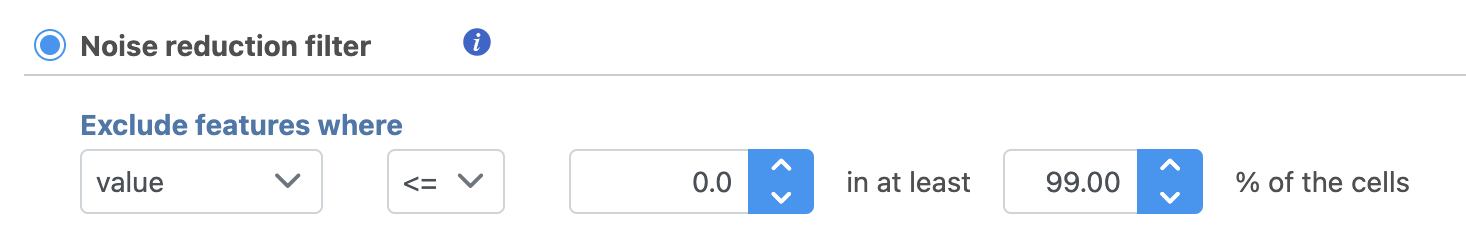
For example, you can use a noise reduction filter to exclude genes that are not expressed by any cell in the data set, but were included in the matrix file.
- Click the Noise reduction filter check box
- Set the Noise reduction filter to Exclude features where value <= 0 in at least 99.9% of cells using the drop-down menus and text boxes
- Click Finish to apply the filter (Figure 3)
This results node, Filtered counts, will be the starting point for the next stage of analysis.
Normalization
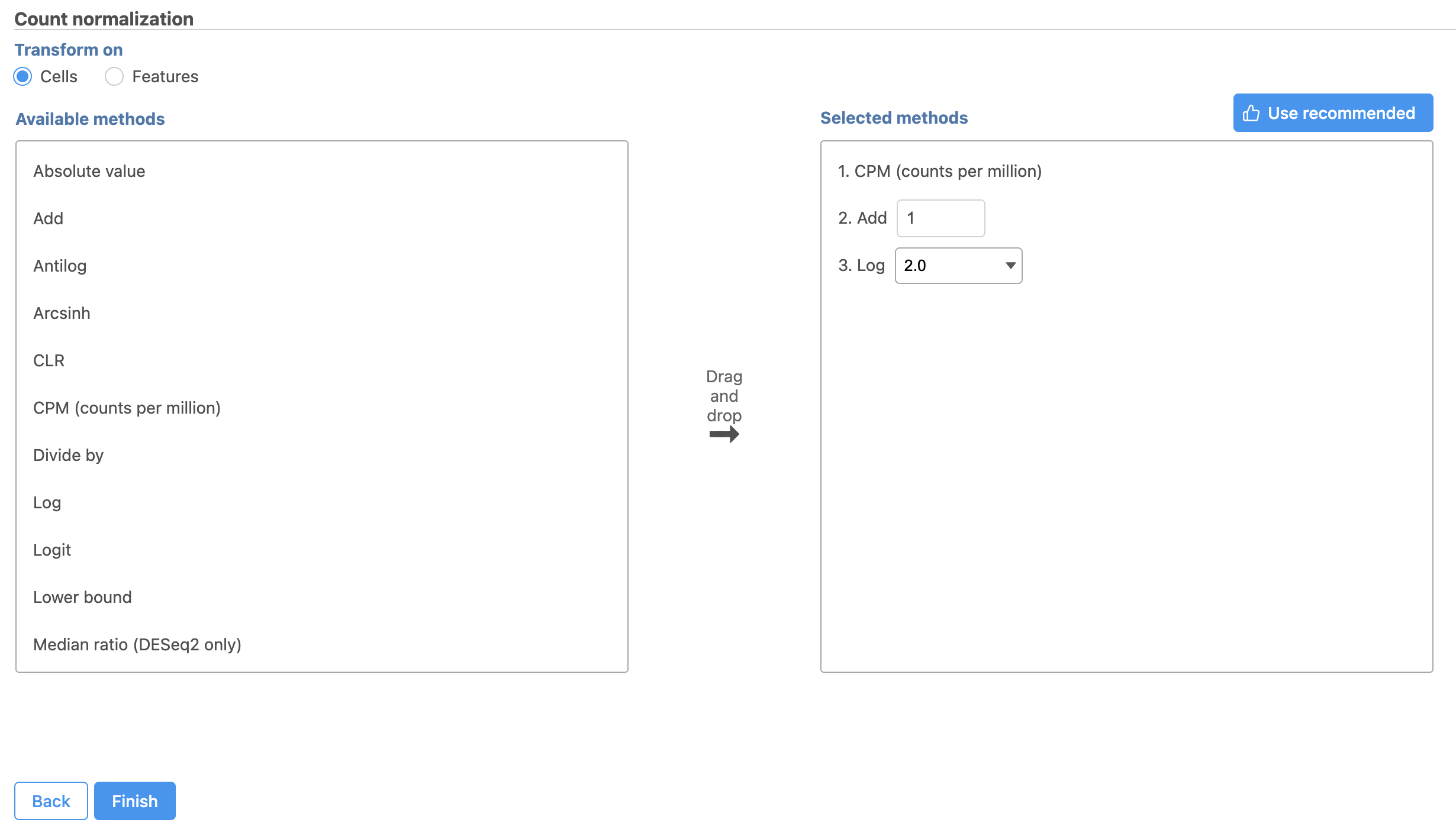
Because different cells will have a different number of total counts, it is important to normalize the data prior to downstream analysis. For droplet-based single cell isolation and library preparation methods that use a 3' counting strategy, where only the 3' end of each transcript is captured and sequenced, we recommend the following normalization - 1. CPM (counts per million), 2. Add 1, 3. Log2. This accounts for differences in total UMI counts per cell and log transforms the data, which makes the data easier to visualize.
- Click the Filtered cells results node produced by the Filtered counts task
- Click Normalization and scaling in the context-sensitive task menu on the right
- Click Normalization
- Click to add the recommended normalization scheme
This adds CPM (counts per million), Add 1, and Log2 to the Normalization order panel. Normalization steps are performed in descending order.
- Click Finish to apply the normalization (Figure 4 )
A new Normalized counts data node will be produced. You can choose to change the color of this node by right-clicking on the task node then clicking Change color and/or rename the result node by right-clicking and selecting Rename data node.
In the example below, I have changed the color to dark blue and renamed the results node based on the scheme.
For more information on normalizing data in Partek Flow, please see the Normalize Counts section of the user manual.
PCA
Principal components (PC) analysis (PCA) is an exploratory technique that is used to describe the structure of high dimensional data by reducing its dimensionality. Because PCA is used to reduce the dimensionality of the data prior to clustering as part of a standard single cell analysis workflow, it is useful to examine the results of PCA for your data set prior to clustering.
- Click the Filtered counts node
- Click Exploratory analysis in the task menu
- Click PCA from the drop-down list
You can choose Features contribute equally to standardize the genes prior to PCA or allow more variable genes to have a larger effect on the PCA by choosing by variance. By default, we take variance into account and focus on the most variable genes.
If you have multiple samples, you can choose to run PCA for each sample individually or for all samples together by selecting or not selecting the Split by sample option (Figure 5).
- Click Finish to run
A new PCA task node will be produced.
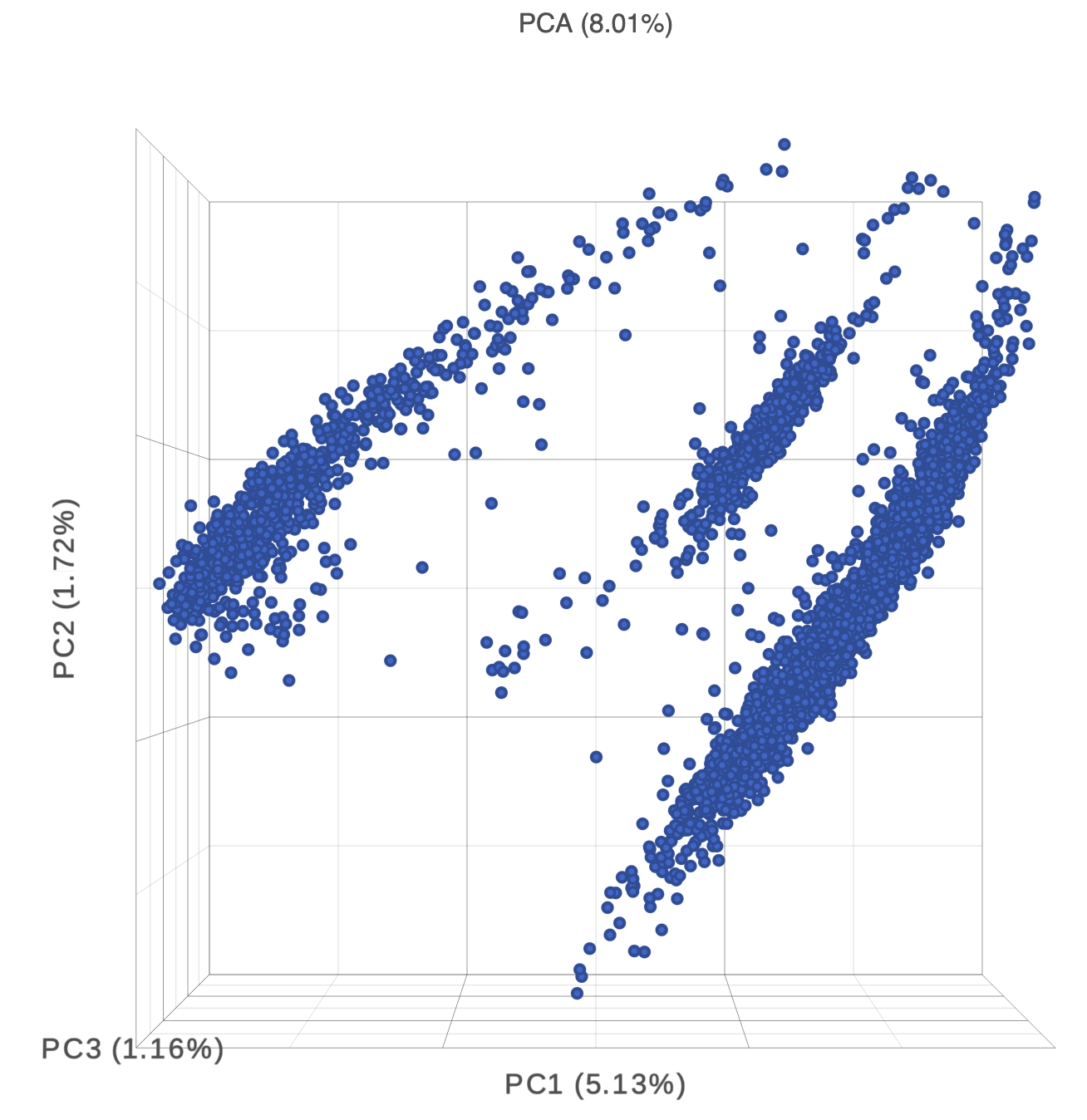
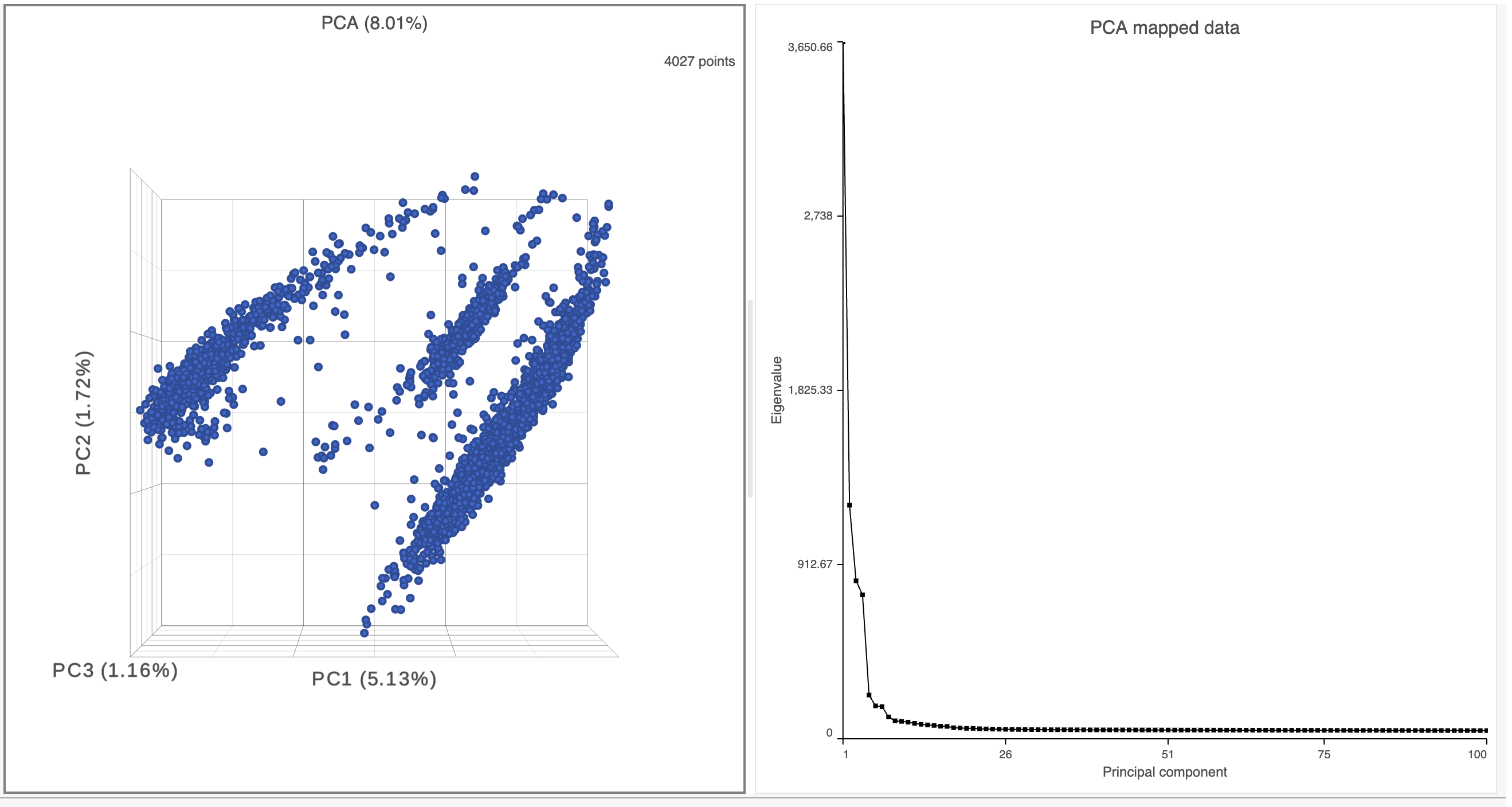
- Double-click the PCA task node to open the 3D PCA scatter plot in data viewer (Figure 6)
The Scree plot lists PCs on the x-axis and the amount of variance explained by each PC on the y-axis, measured in Eigenvalue. The higher the Eigenvalue, the more variance is explained by the PC. Typically, after an initial set of highly informative PCs, the amount of variance explained by analyzing additional PCs is minimal. By identifying the point where the Scree plot levels off, you can choose an optimal number of PCs to use in downstream analysis steps like graph-based clustering, UMAP and t-SNE.
- To draw a Scree plot, in Data viewer, choose Scree plot icon available in New plot under Setup on the left panel , choose the PCA data node (Figure 7)
Note that Partek Flow suggests appropriate data for each plot type that is chosen so only PCA results will be available to select from for the Scree plot.
- Mouse over the Scree plot to identify the point where additional PCs offer little additional information
In this data set, a reasonable cut-off could be set anywhere between 7 and 20 PCs.
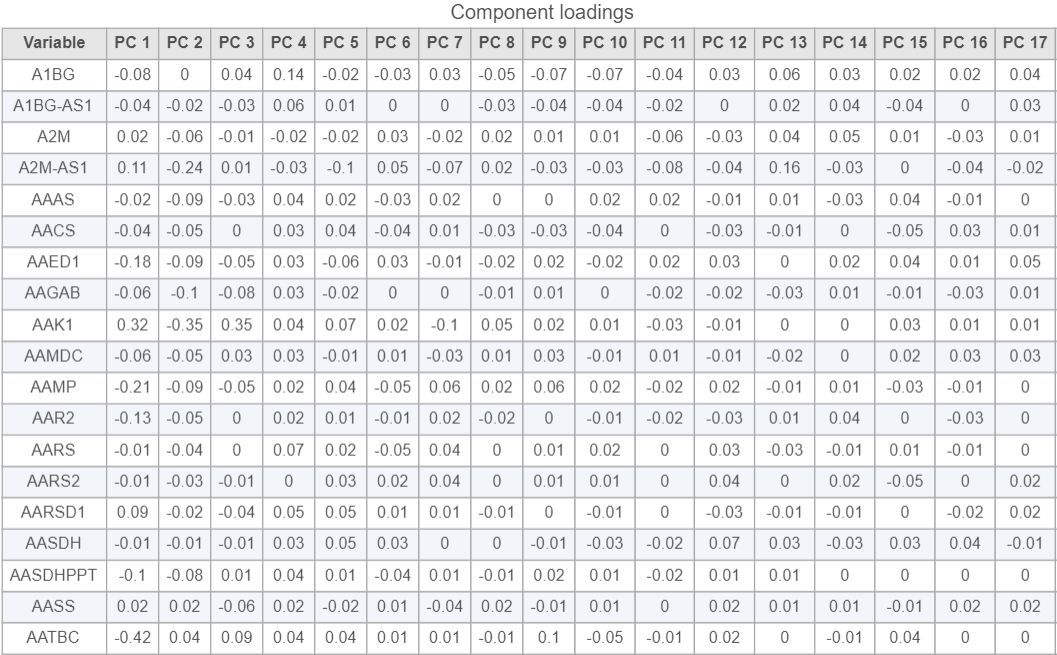
Viewing the genes correlated with each PC can be useful when choosing how many PCs to include.
- Click the Table option in the New plot icon under Setup and select the PCA data node to open the Component loadings table (Figure 8)
To display PCA projects table, click on the Table drop-down list in the Content icon under Configure and choose PCA projections (Figure 9)
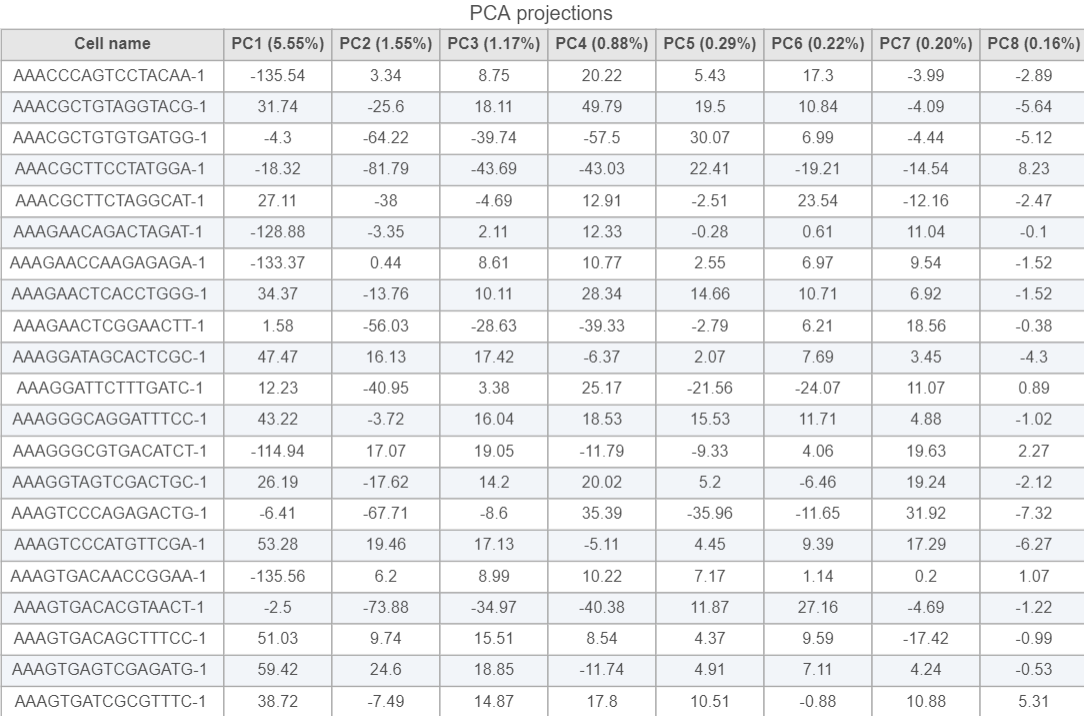
PCA projections table contains each row as an observation (a cell in this case), each column represents one principal component (Figure 10). This table can be downloaded as text file, the same way as the component loading table.
Graph-based clustering
Graph-based clustering identifies groups of similar cells using PC values as the input. By including only the most informative PCs, noise in the data set is excluded, improving the results of clustering.
- Click the PCA data node
- Click Exploratory analysis in the task menu
- Click Graph-based clustering
Clustering can be performed on each sample individually or on all samples together. Here, we are working with a single sample.
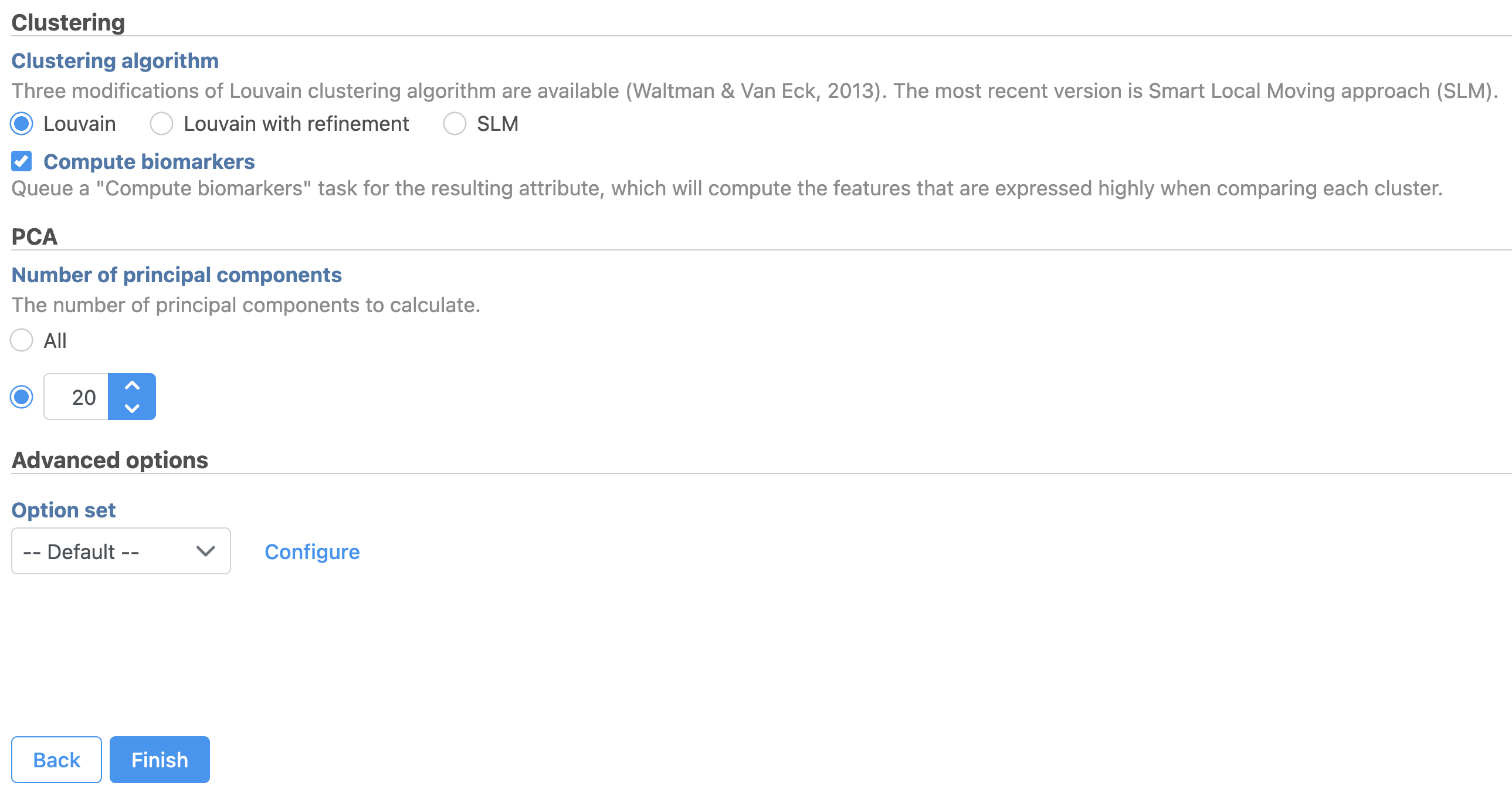
- Check Compute biomarkers to compute features that are highly expressed when comparing each cluster (Figure 11)
- Click Configure to access the Advanced options and change the Number of nearest neighbors to 50 and Nearest Neighbor Type to K-NN for this example tutorial.
The Number of principal components should be set based on the your examination of the Scree plot and component loadings table. The default value of 100 is likely exhaustive for most data sets, but may introduce noise that reduces the number of clusters that can be distinguished.
- Click Finish to run the task
A new Graph-based clusters data and Biomarkers data node will be generated along with the task nodes.
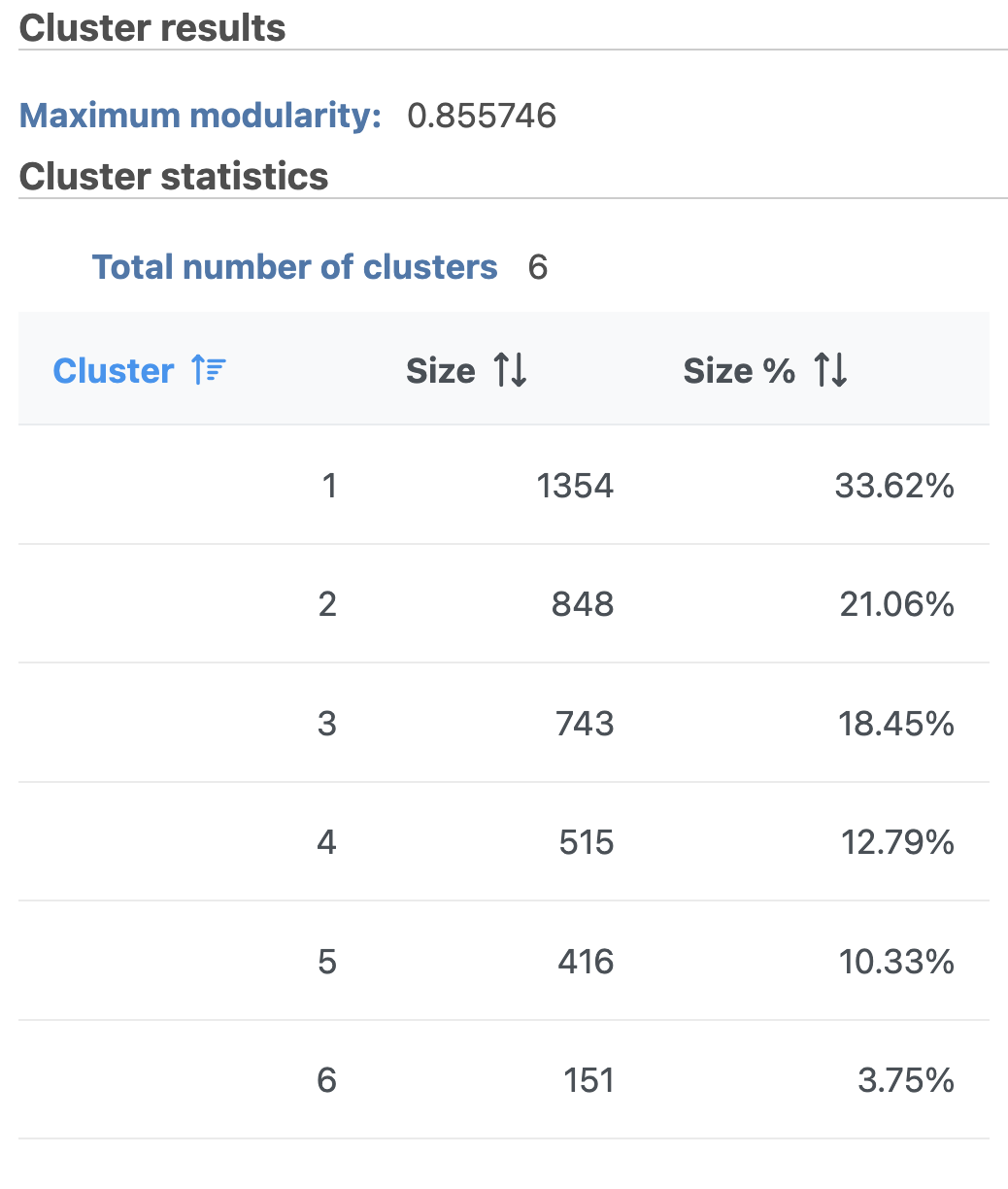
- Double-click the Graph-based clusters node to see the cluster results and statistics (left screenshot on Figure 12)
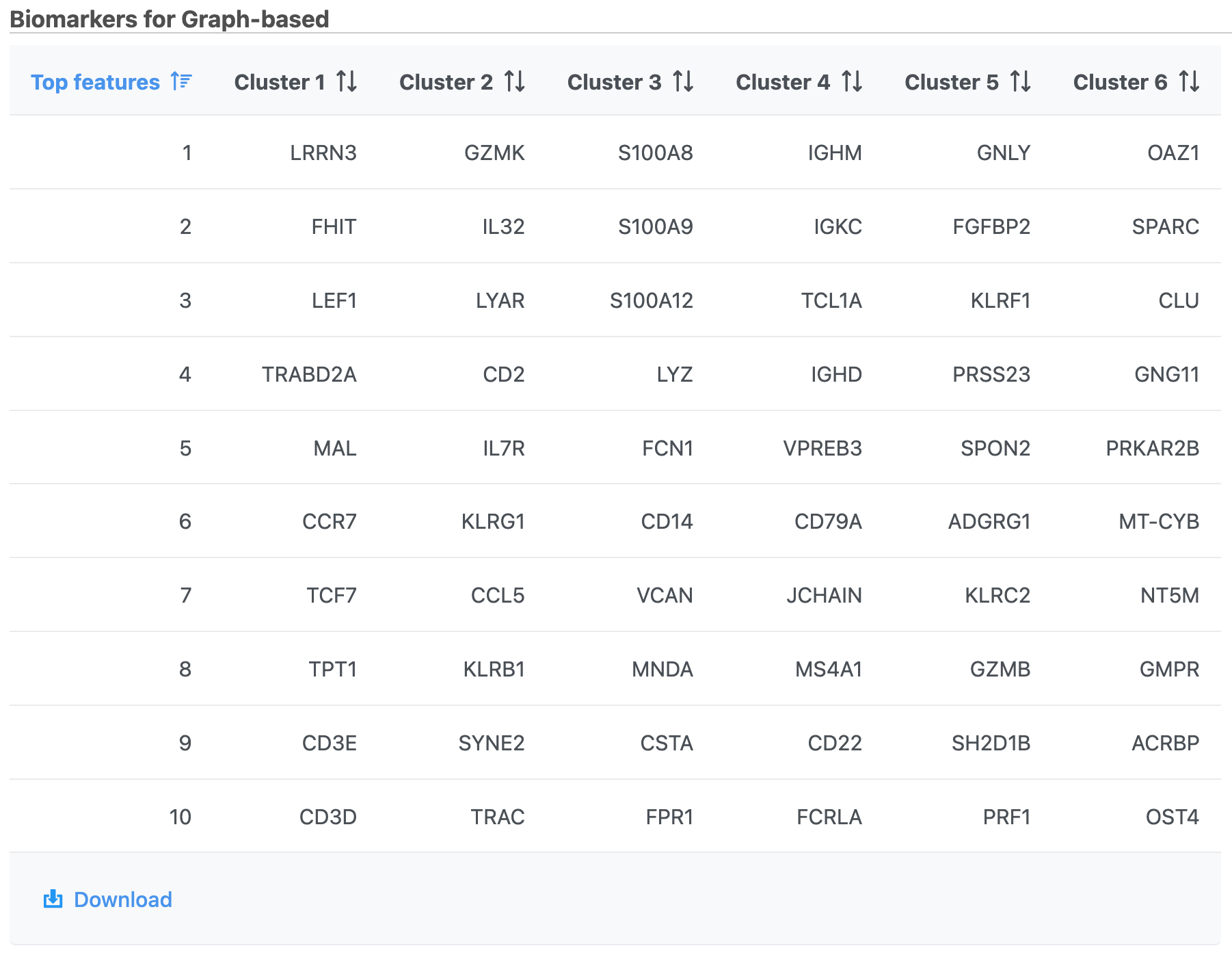
- Double-click the Biomarkers node to see the computed biomarkers if you have selected this option (right screenshot on Figure 12)
The Graph-based clustering result lists the Total number of clusters and what proportion of cells fall into each cluster as well as Maximum modularity which is a measurement of the quality of the clustering result. The Biomarkers node includes the top features for each graph-based cluster. It displays the top-10 genes that distinguish each cluster from the others. Download at the bottom right of the table can be used to view and save more features. These are calculated using an ANOVA test comparing the cells in each group to all the other cells, filtering to genes that are 1.5 fold upregulated, and sorting by ascending p-value. This ensures that the top-10 genes of each cluster are highly and disproportionately expressed in that cluster.
t-SNE
t-Distributed Stochastic Neighbor Embedding (t-SNE) is a dimensional reduction technique that prioritizes local relationships to build a low-dimensional representation of the high-dimensional data that places objects that are similar in high-dimensional space close together in the low-dimensional representation. This makes t-SNE well suited for analyzing high-dimensional data when the goal is to identify groups of similar objects, such as cell types in single cell RNA-Seq data.
- Click the Graph-based clusters node
- Click Exploratory analysis in the task menu
- Click t-SNE
If you have multiple samples, you can choose to run t-SNE for each sample individually or for all samples together using the Split cells by sample option. Please note that this option will not be present if you are running t-SNE on a clustering result. For clarity, clustering results run with all samples together must be viewed together and clustering results run by sample must be viewed by sample.
Like Graph-based clustering, t-SNE takes PC values as its input and further reduces the data down to two or three dimensions. For consistency, you should use the same number of PCs as the input for t-SNE that you used for Graph-based clustering.
- Click Apply
- Click Finish to run (Figure 13)
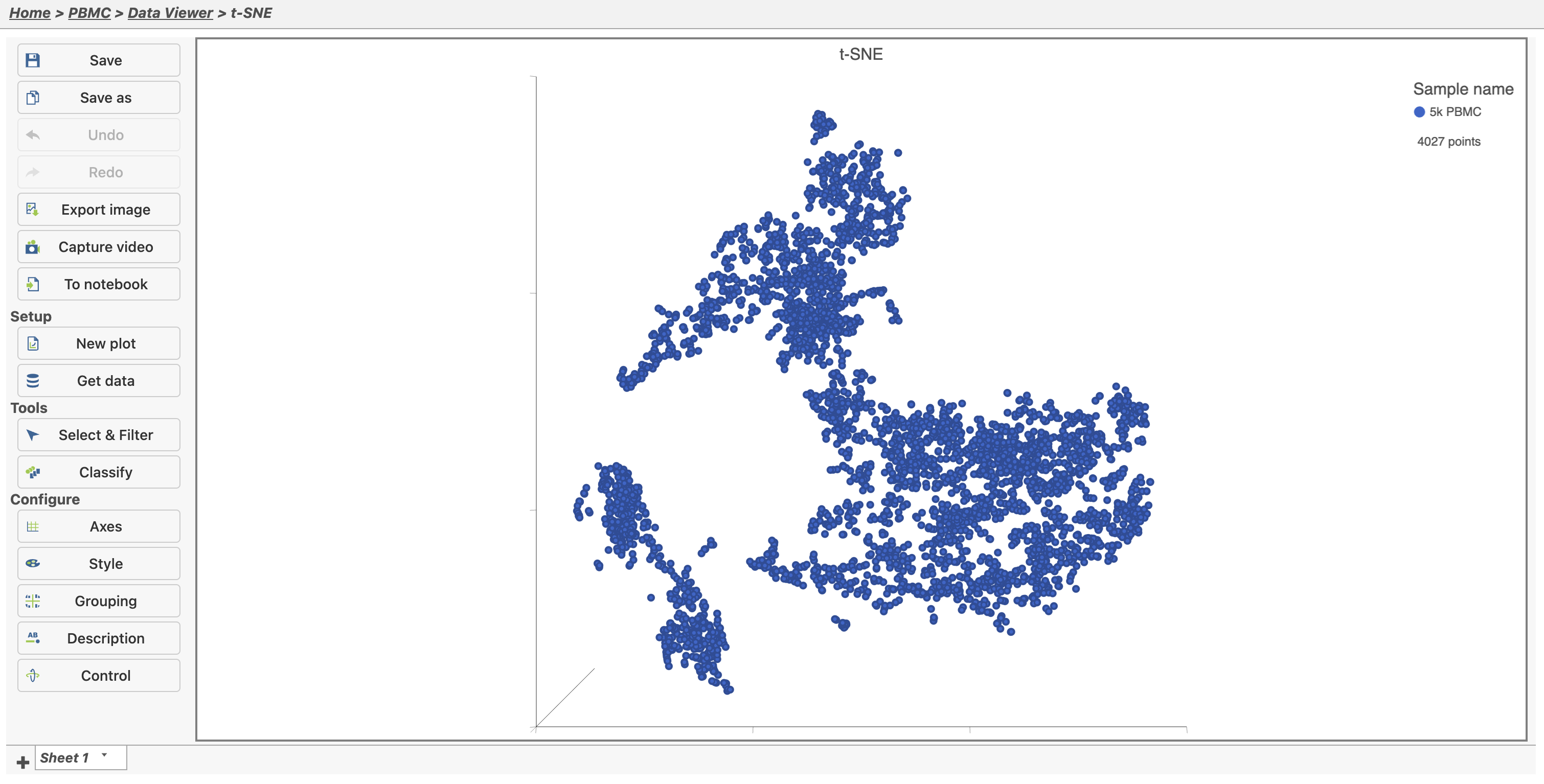
- Double-click the t-SNE node to open the t-SNE task report (Figure 14). Use the panel on the left to modify the plot or add more plots to this Data viewer session.
Coloring the t-SNE scatter plot
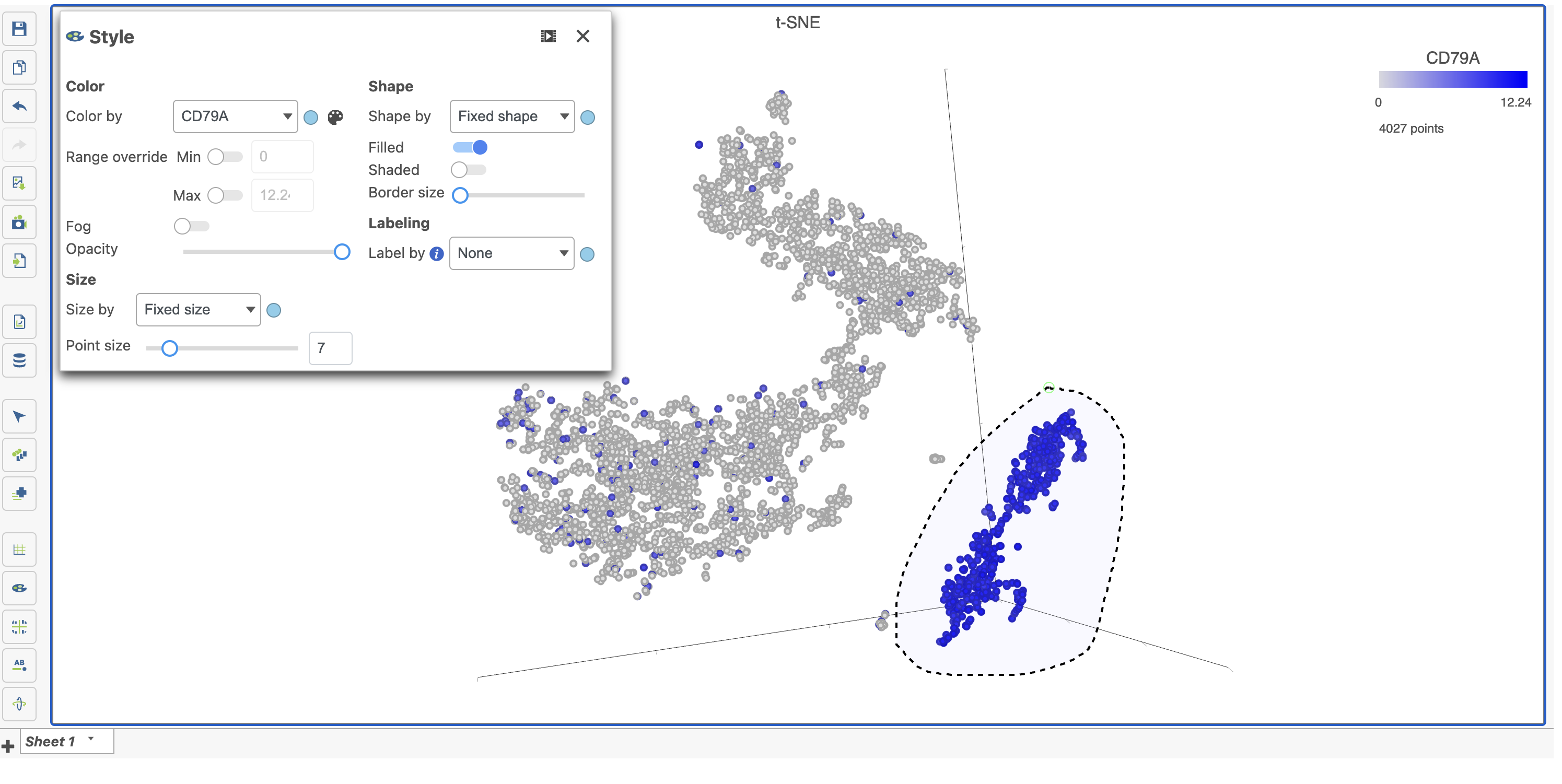
Click on the plot to ensure that the plot window is selected. Click Style under Configure to color the t-SNE.
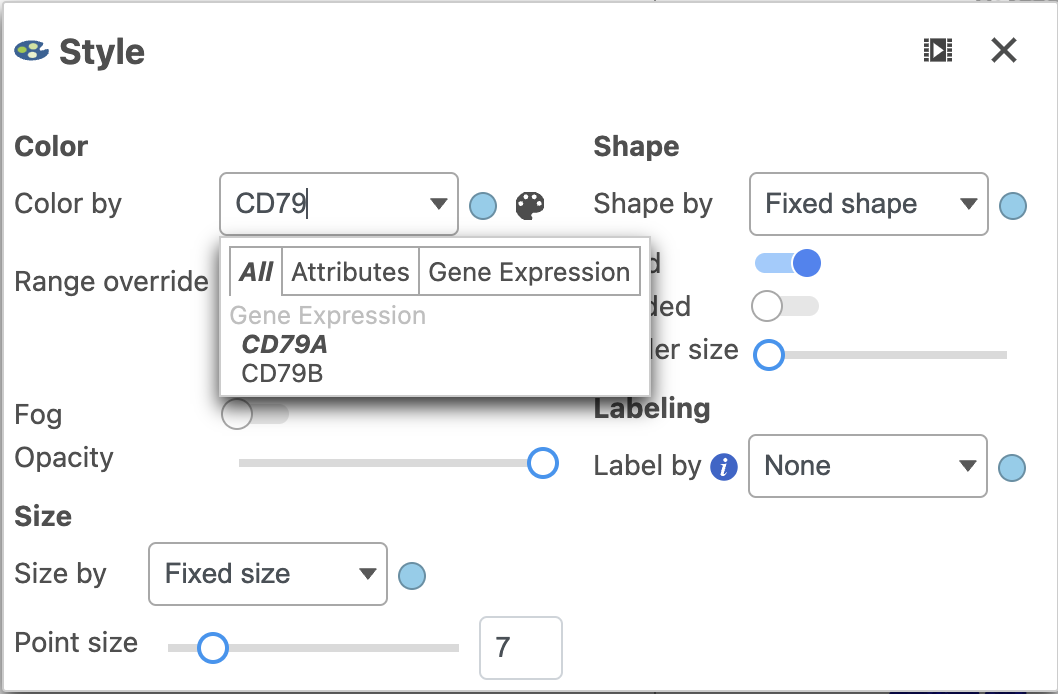
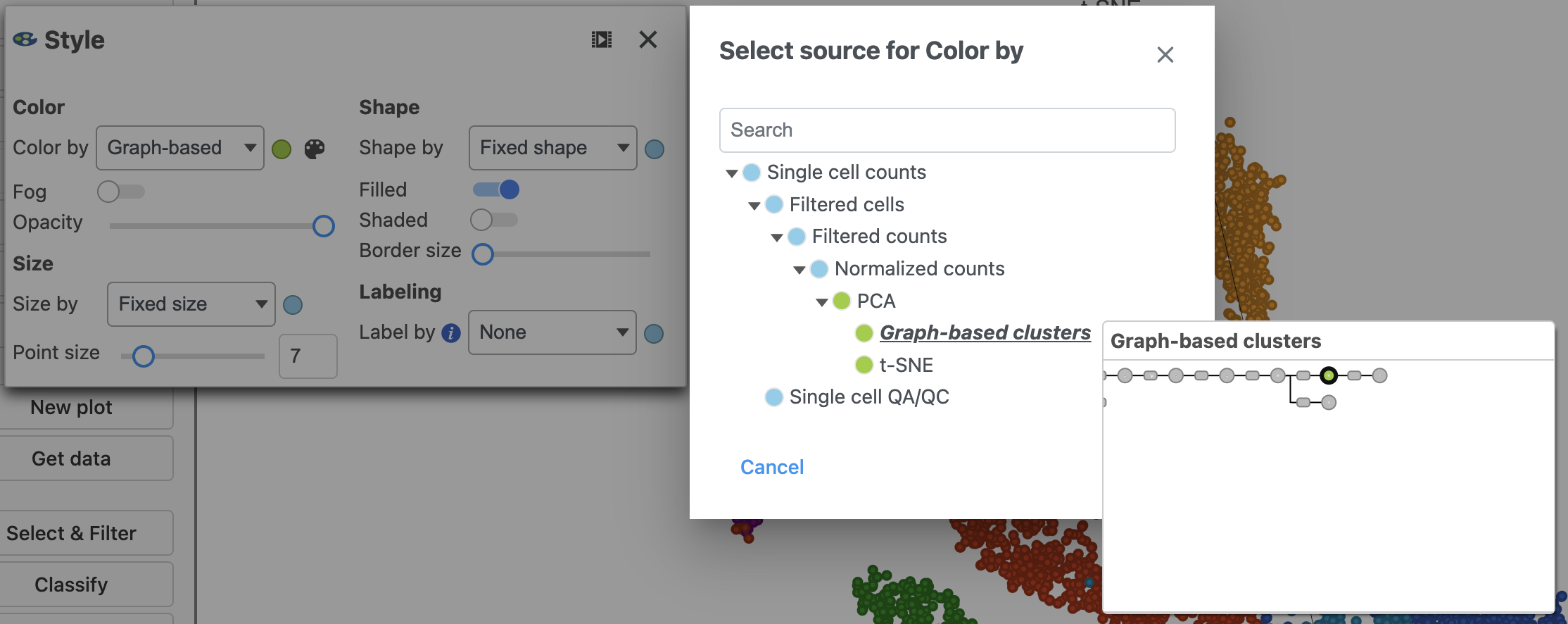
- Color by the options in the drop-down menu under Color. You should be on the normalized counts node which can be seen by hovering over or clicking the circle (node) to the right of the drop-down.
- Click the text field in the drop-down and start typing CD79A then select the gene by clicking on it (Figure 15)
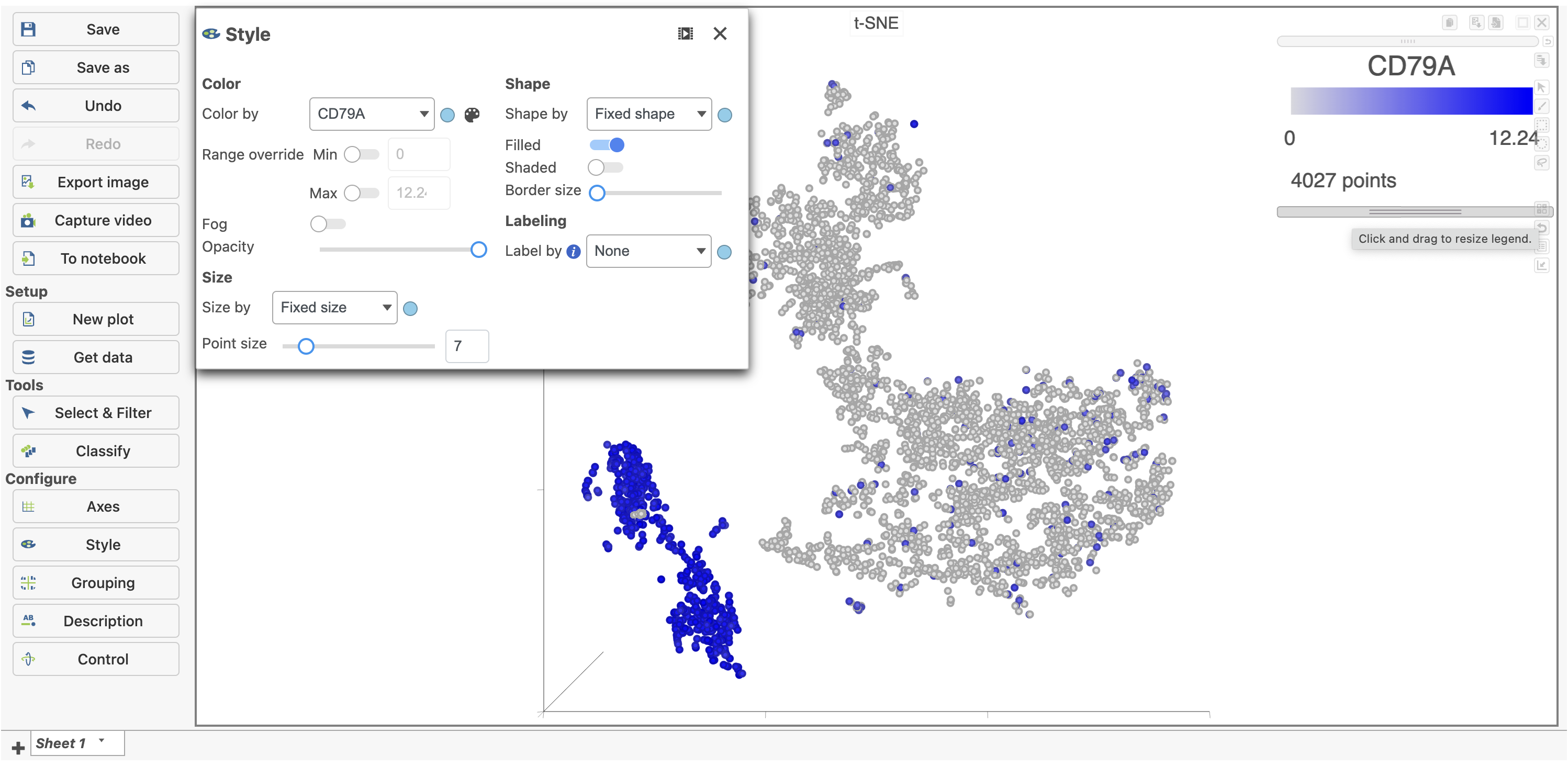
The cells on the plot will be colored based on their expression level of CD79A (Figure 16). In the example in Figure 16, the Style icon has been dragged to a different location on the screen and the legend has also been resized and moved. Resizing the legend can either be done on the legend itself or using the Description icon under Configure.
Clicking a cell on the plot shows the expression values of the cell in the legend. Hovering over a cell on the plot also shows this information and related details (Figure 18).
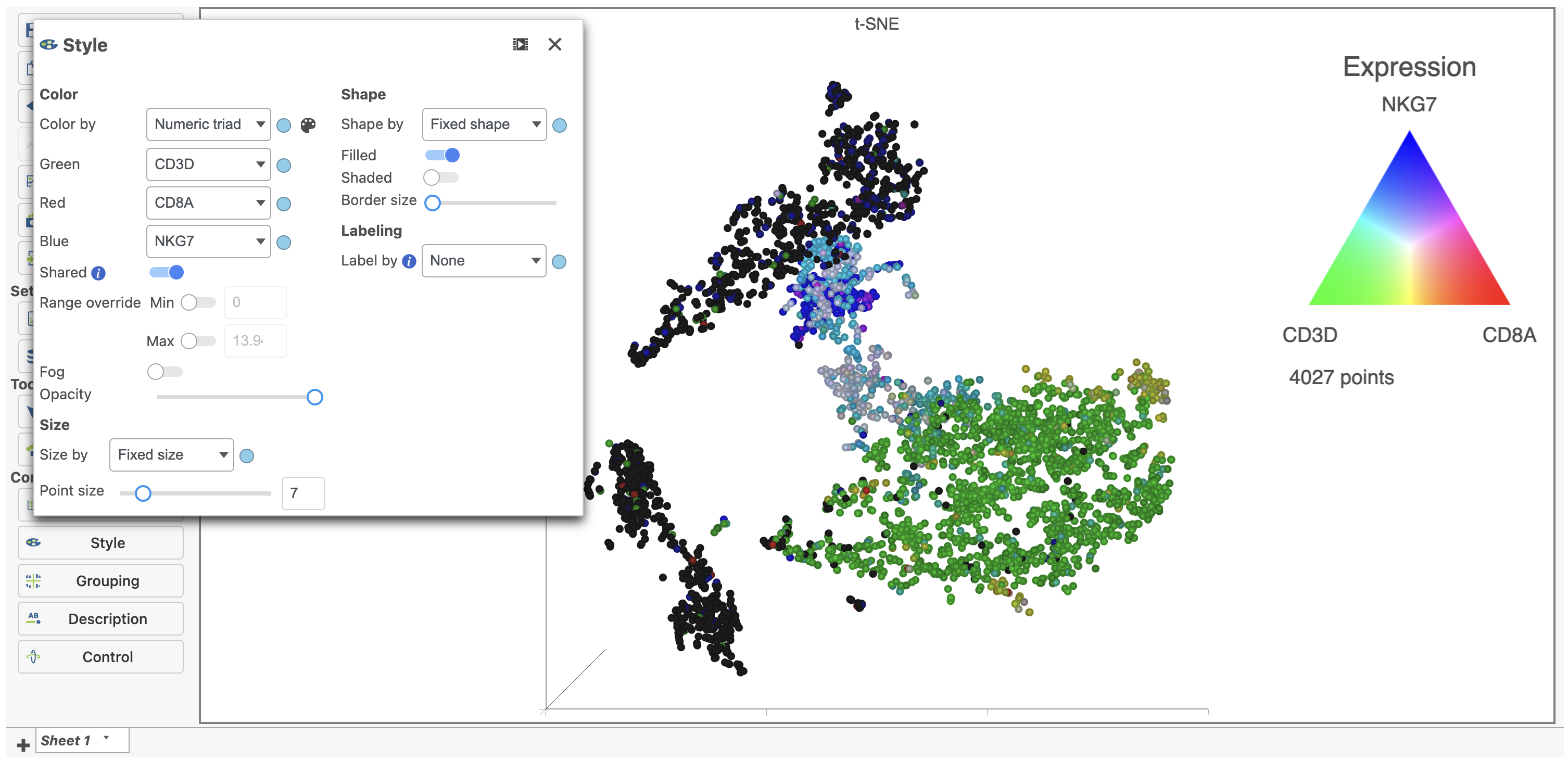
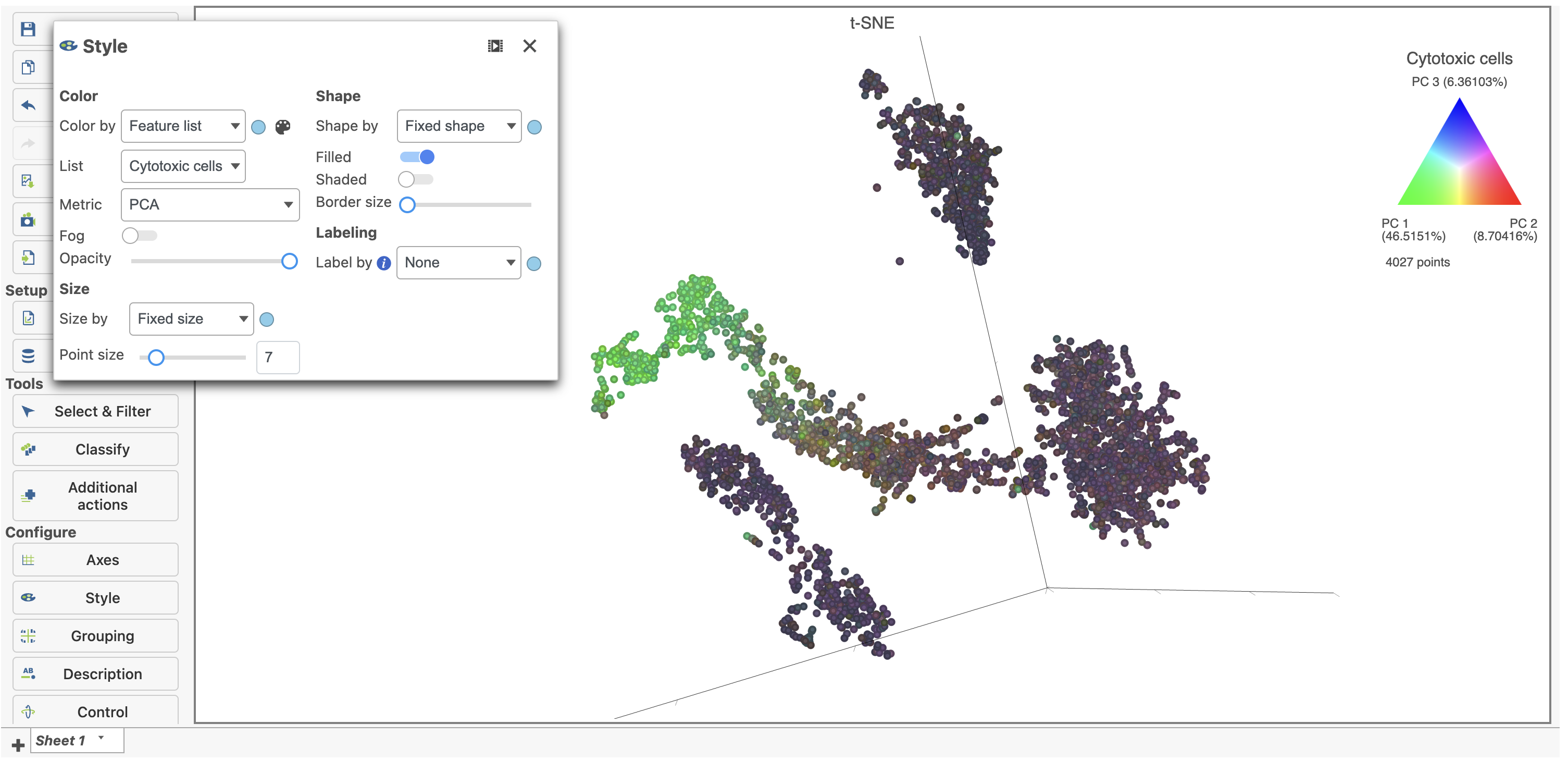
If you want to color by more than three genes at time, such as by a list of genes that distinguish a particular cell type, you can use the color by Feature list option.
- Select Feature List from the Color by drop-down
- Choose Cytotoxic cells from the List drop-down (use List management in Settings to add lists to Partek Flow which will automatically make them available here)
- Choose PCA from the Metric drop-down
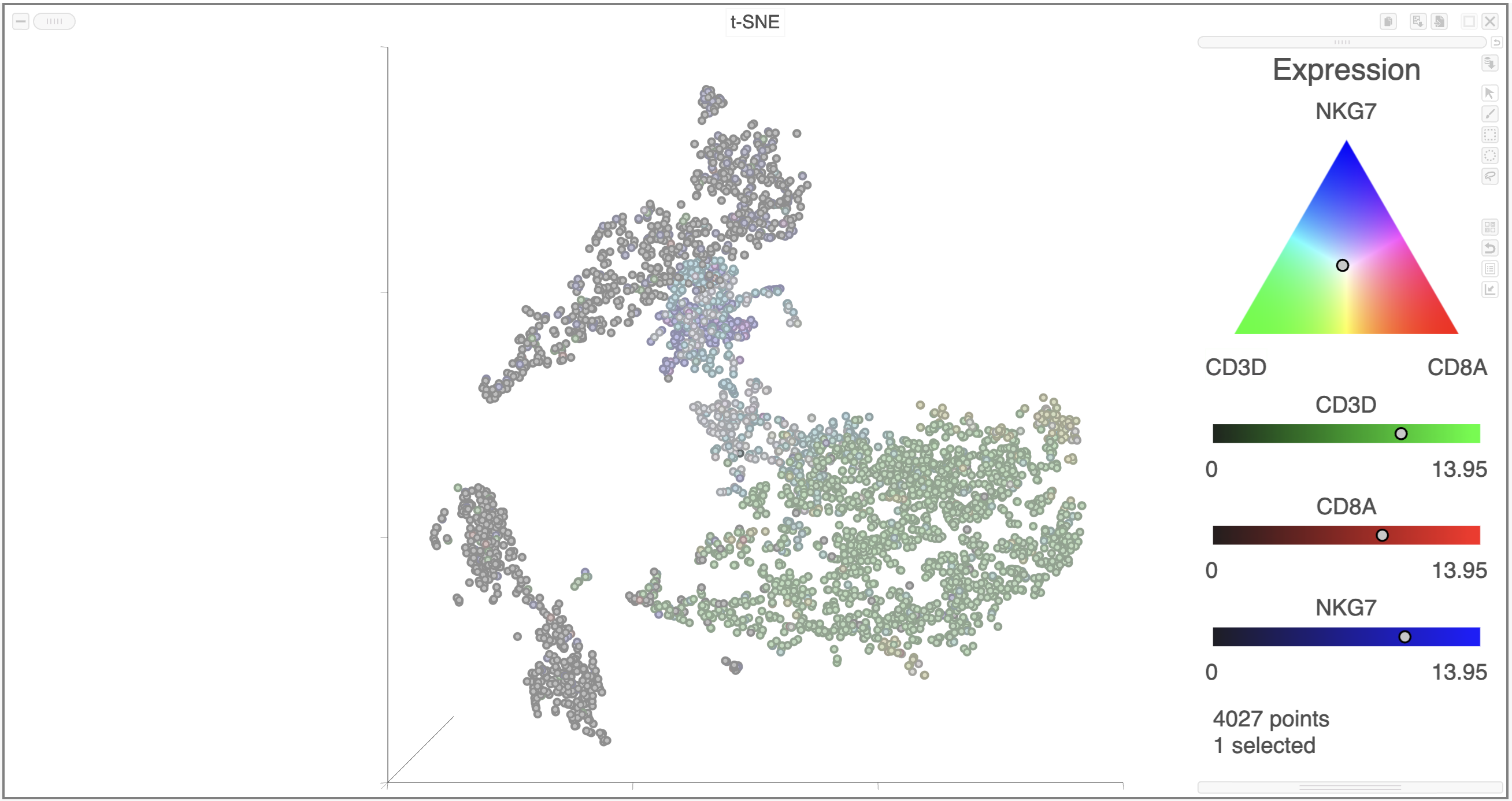
Coloring by a list, in this way, calculates the first three principal components for the gene list and colors the cells on the plot by their values along those three PCs with green for PC1, red for PC2, and blue for PC3 (Figure 19).
In addition to coloring by gene expression and by gene lists, the points can be colored by any cell or sample attribute. Available attributes are listed as options in the Color by drop-down menu. Note that any available options are dependent upon the selected data node. In the following section we will use the attribute Graph-based to color our cells by the clusters identified in the Graph-based clustering task (Figure 20).
Selecting cells on the t-SNE scatter plot
The most basic way to select a point on the scatter plot is to click it with the mouse while in pointer mode. To select multiple cells, you can hold Ctrl on your keyboard and click the cells. To select larger groups of cells, you can switch to Lasso mode by clicking in the plot controls on the right hand side. The lasso lets you freely draw a shape to select a cluster of cells.
- Click to activate Lasso mode
- Left-click and hold to draw a lasso around a cluster of cells
- Release and click the starting circle to close the lasso and select the enclosed cells (Figure 21)
You can also create a lasso with straight lines using Lasso mode by clicking, releasing, and clicking again to draw a shape.
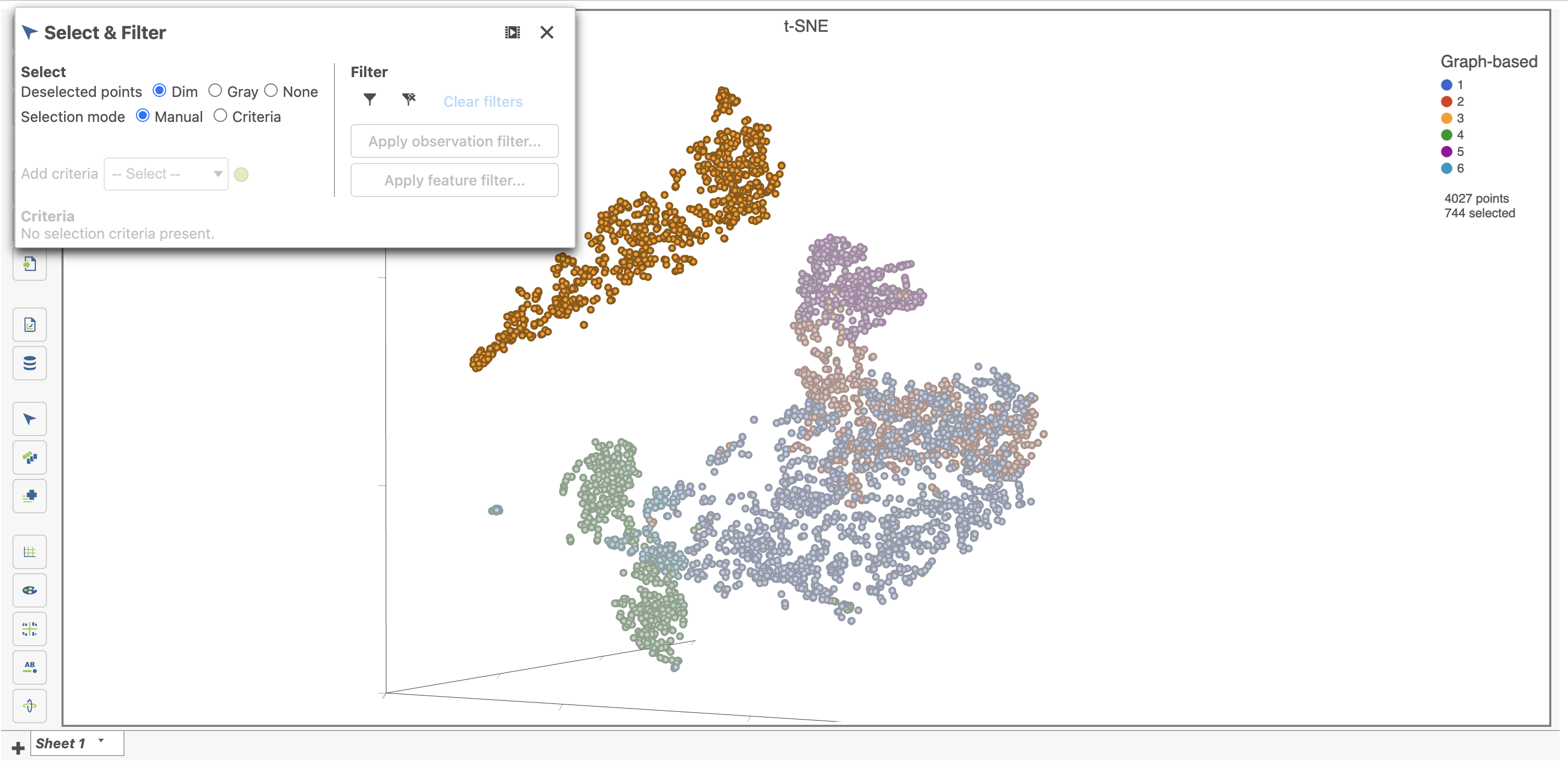
By default, selected cells are shown in bold while unselected cells are dimmed (Figure 22). This can be changed to gray selected cells using the Select & Filter tool in the left panel as shown in Figure 22.
- Double-click any blank section of the scatter plot to clear the selection
Alternatively, you can select cells using any criteria available for the data node that is selected in the Select & Filter tool. To change the data selection click the circle (node) and select the data.
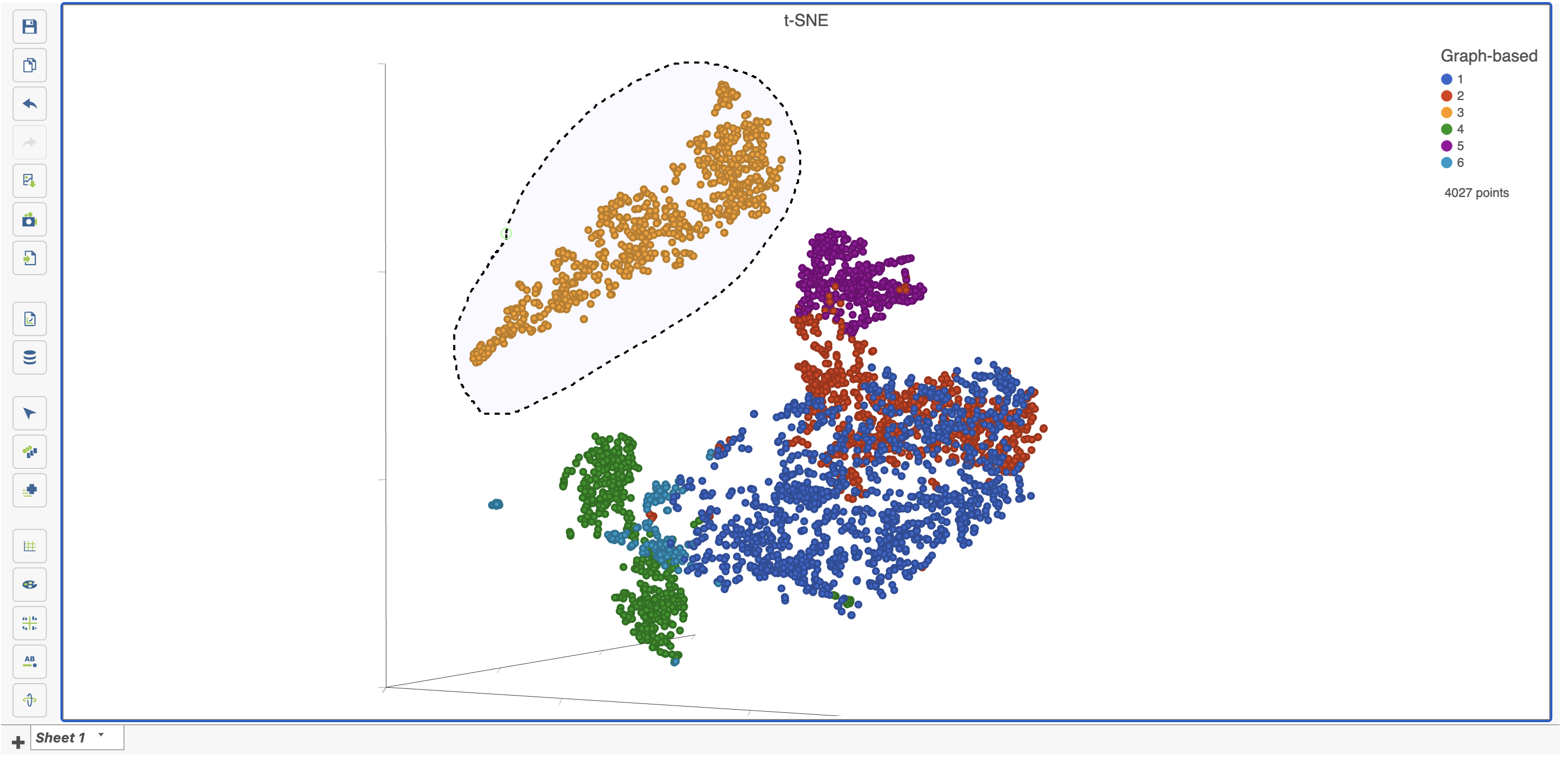
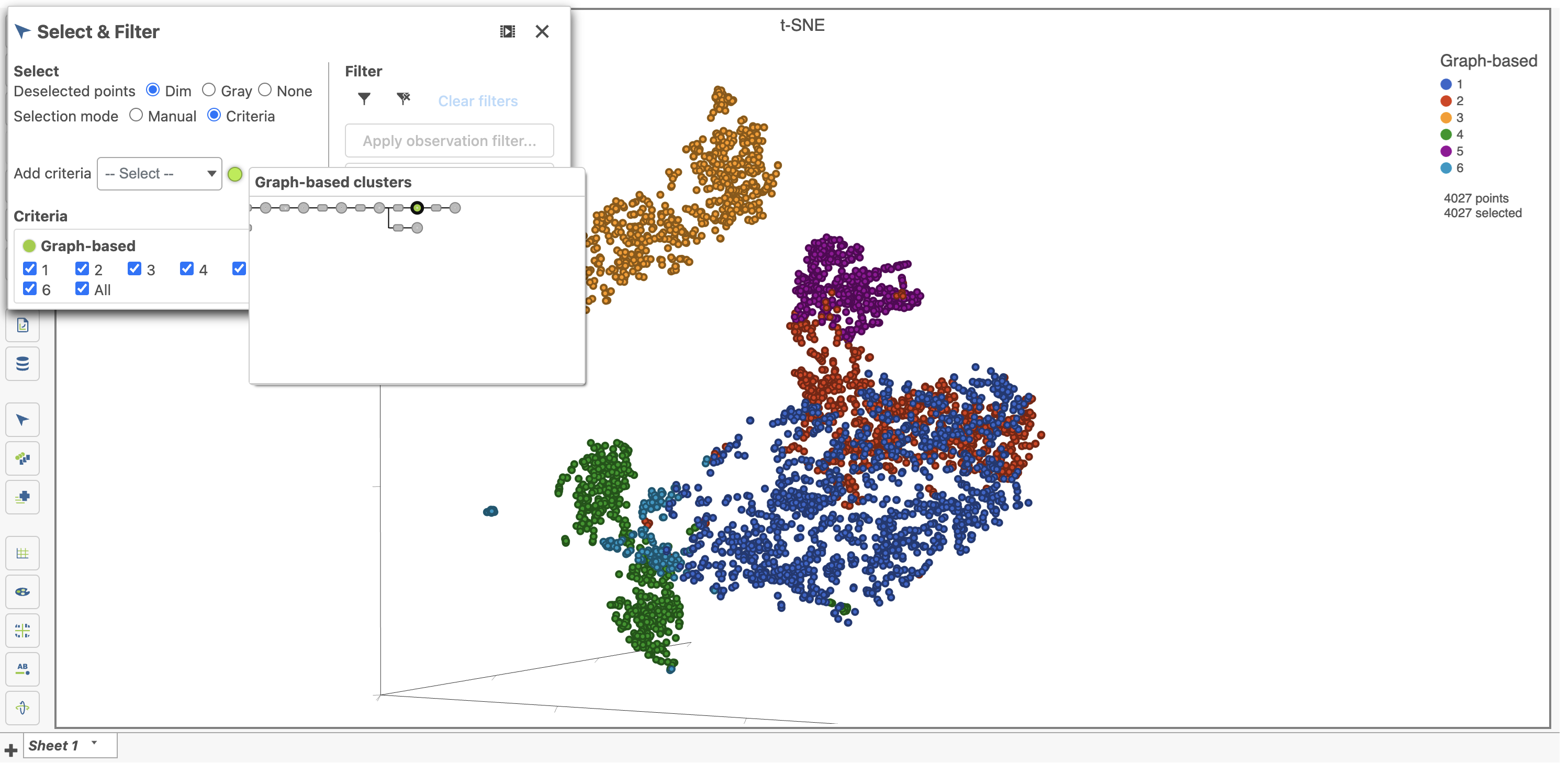
- Choose Graph-based from the Criteria drop-down menu in the Select & Filter tool after ensuring you on are on the Graph-based cluster node by hovering on the circle (Figure 23). If you are not on the correct node, you need to click the circle and select the data.
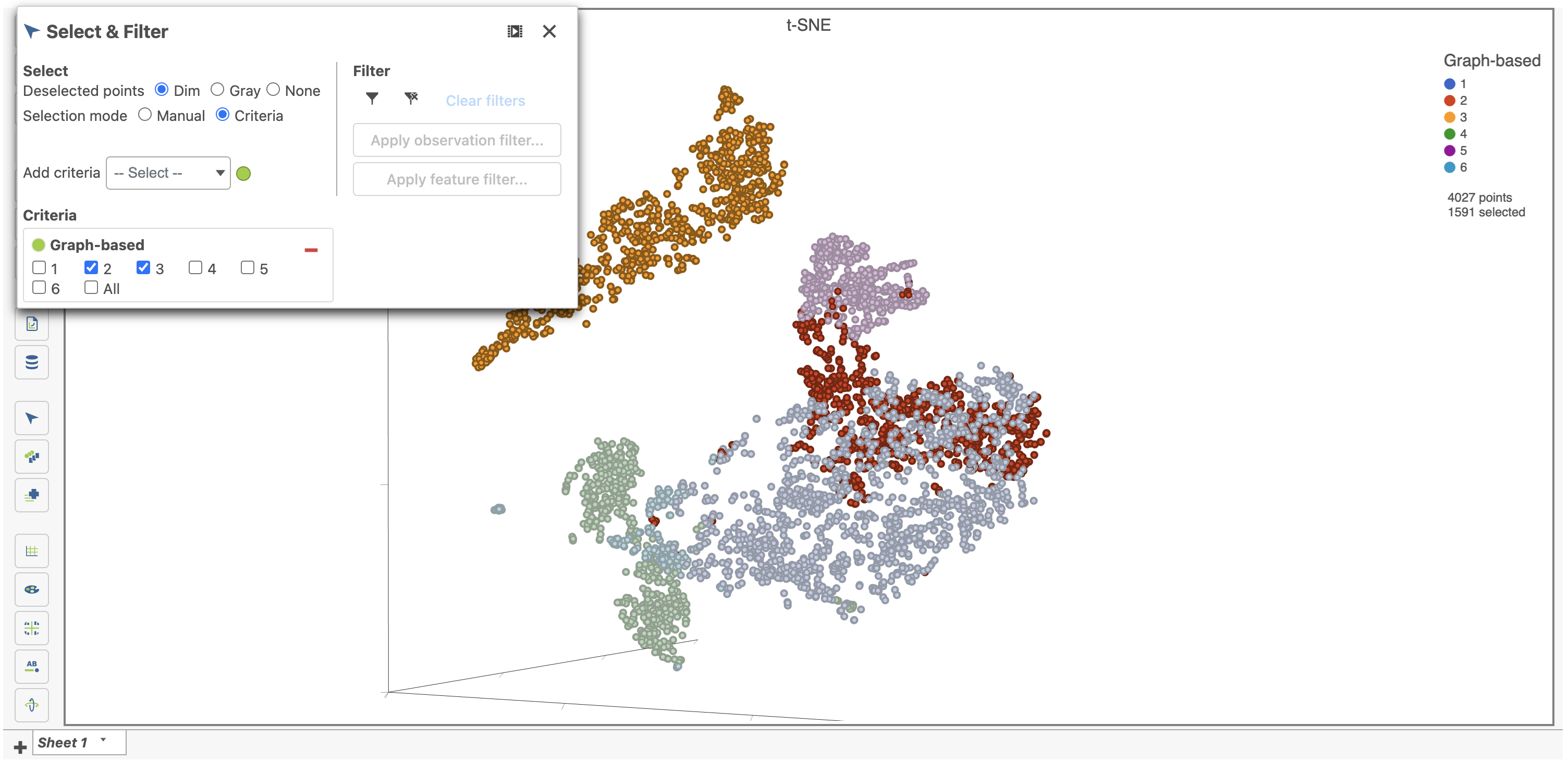
- Click only 2 and 3
This selects cells from Graph-based clusters 2 and 3 (Figure 24). The number of selected cells is listed in the Legend on the plot.
Cells can also be selected based on their gene expression values in the Select & Filter section.
- Click the circle and select the Normalized counts node which has gene expression data
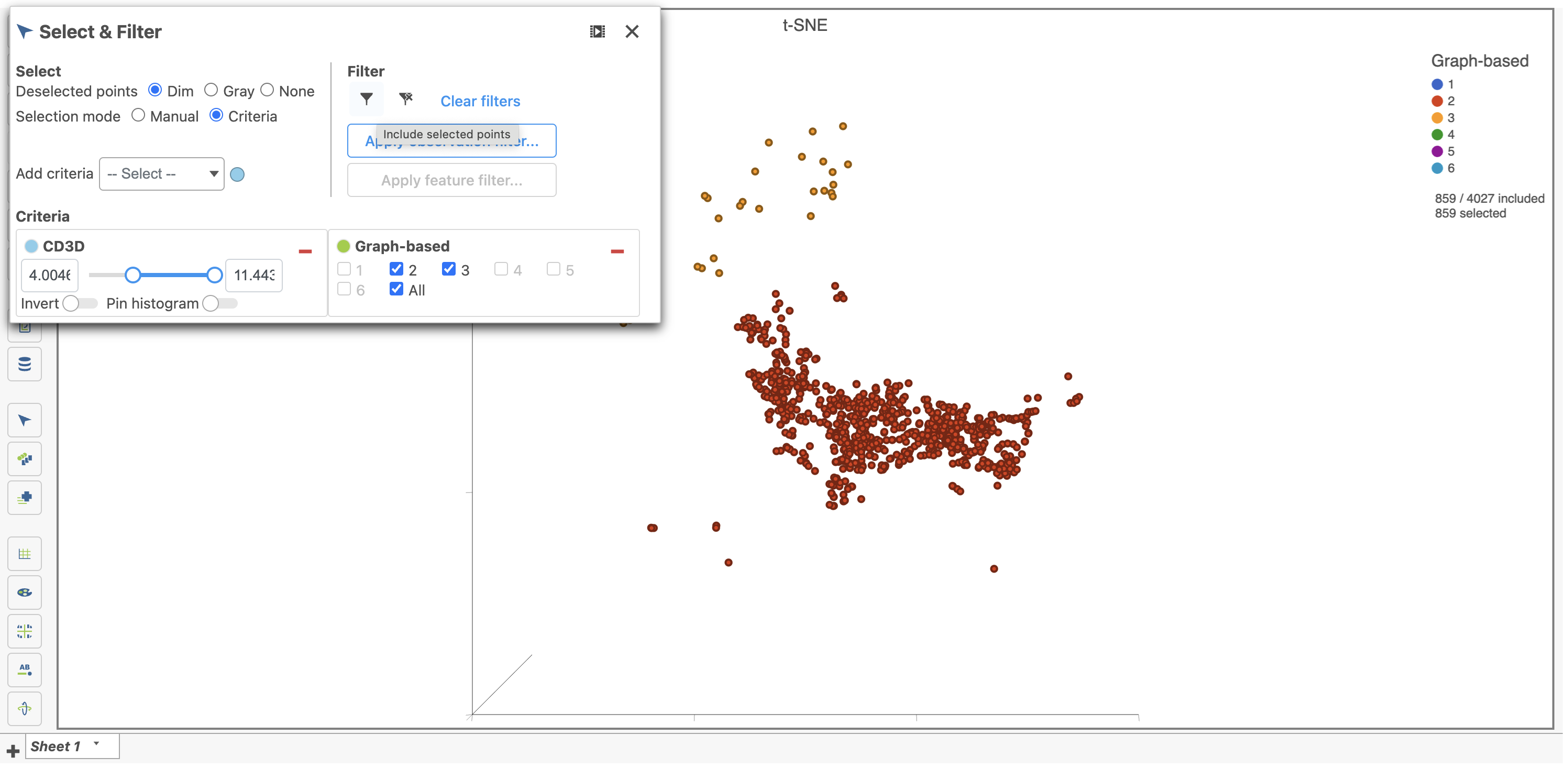
- Type cd3d in the text field of the drop-down
- Click on CD3D to add it as criteria to select from and use the slider or text field to adjust the selected values. Pin the histogram to visualize the distribution during selection.
Very specific selections can be configured by adding criteria in this way. In the example below, Clusters 2 and 3 and high CD3D expression is selected (Figure 25).
Filtering cells on the t-SNE scatter plot
Once a cell has been selected on the plot, it can be filtered. The filter controls can exclude or include (only) any selected cell. Filtering can be particularly useful when you want to use a gene expression threshold to classify a group of cells, but the gene in question is not exclusively expressed by your cell type of interest.
In this example we can filter to include just cells from the selection we have already made.
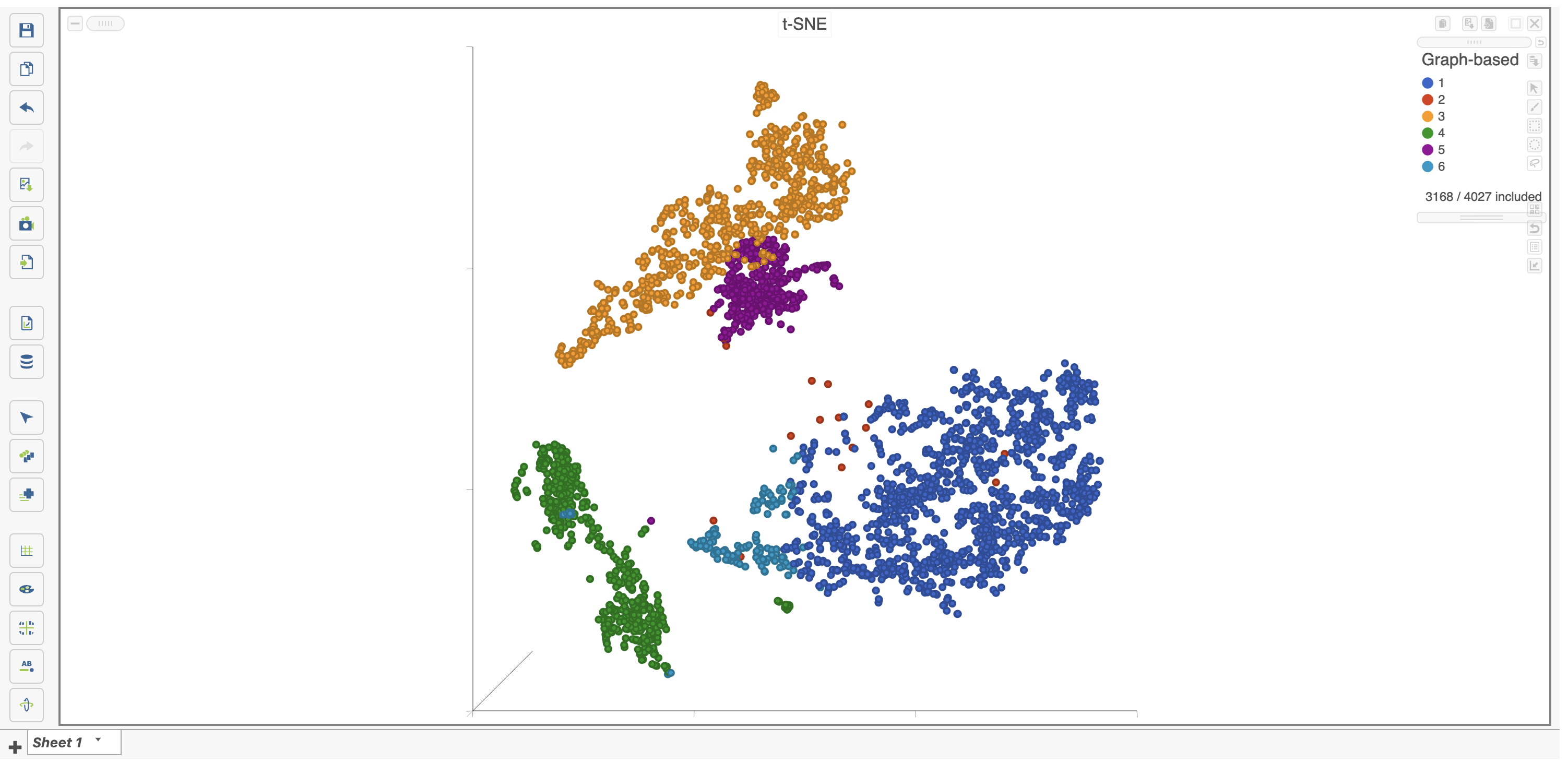
- Click (filter include) to filter to just the selected cells (Figure 26).
The plot will update to show only the included cells as seen in Figure 26.
Cells that are not shown on the plot cannot be selected, allowing you to focus on the visible cells. The number of cells shown on the plot out of the total number of original cells is shown in the Legend. You can adjust the view to focus on only the included cells.
- Click on the plot controls or toggle on Fit visible in the Axes configuration to rescale the axes to the filtered points
To revert to the original scaling, click the button again or turn off Fit visible with the toggle.
- Alternatively, to exclude selected cells, click (filter exclude) (Figure 27)
Additional inclusion or exclusion filters can be added to focus on a smaller subset of cells.
- Click Clear filters to remove applied filters
The plot will update to show all cells and return to the original scaling.
Classifying cells
Classifying cells allows to you assign cells to groups that can be used in downstream analysis and visualizations. Commonly, this is used to describe cell types, such as B cells and T cells, but can be used to describe any group of cells that you want to consider together in your analysis, such as cycling cells or CD14 high expressing cells. Each cell can only belong to one class at a time so you cannot create overlapping classes.
To classify a cell, just select it then click Classify selection in the Classify tool.
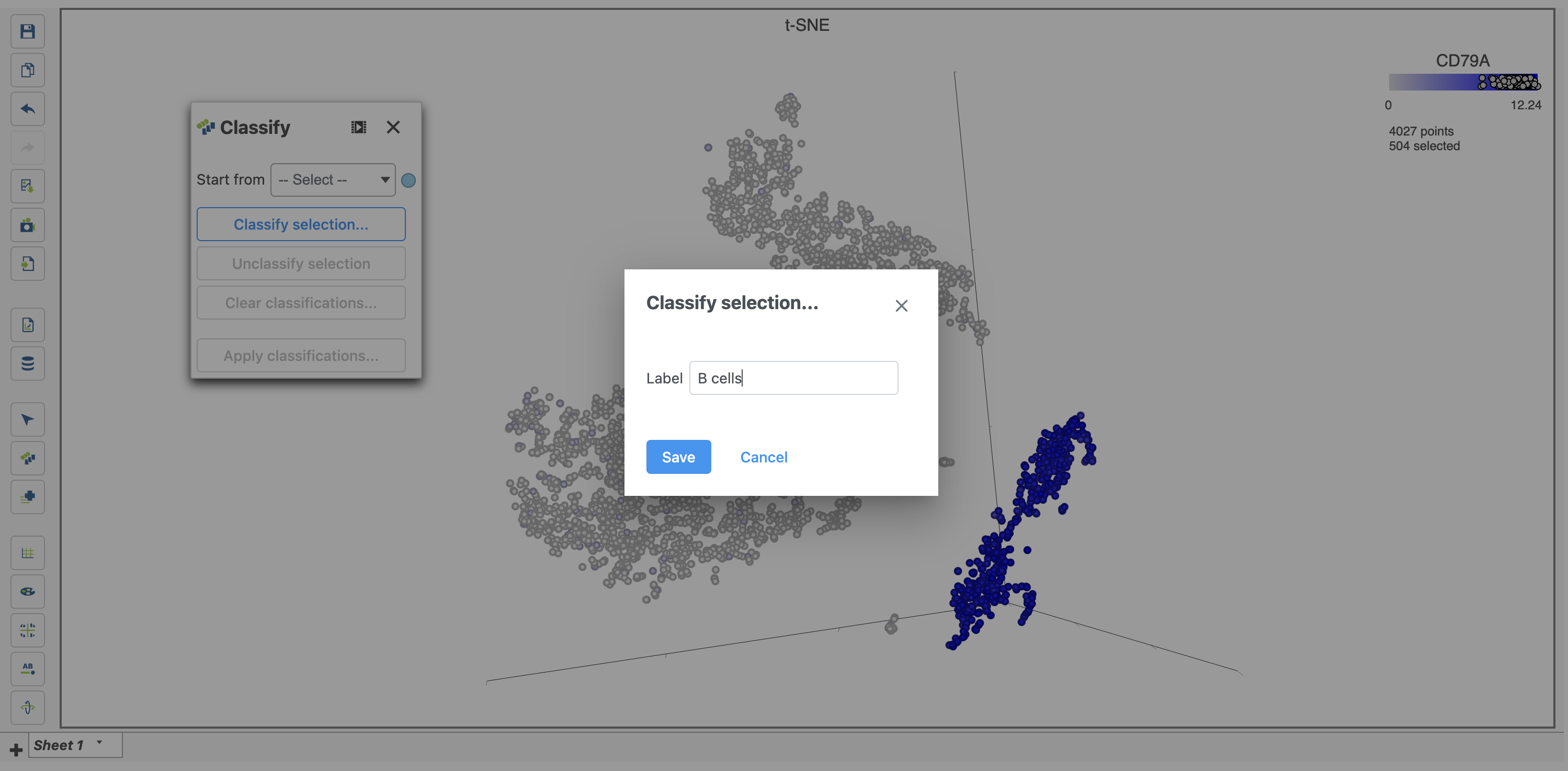
For example, we can classify a cluster of cells expressing high levels of CD79A as B cells.
- Set Color by in the Style configuration to the normalized counts node
- Type CD79A in the search box and select it. Rotate the 3D plot if you need to see this cluster more clearly.
- Click to activate Lasso mode
- Draw a lasso around the cluster of CD79A-expressing cells (Figure 28)
Because most of these cells express CD79A, a B cell marker, and because they cluster together on the t-SNE, suggesting they have similar overall gene expression, we believe that all these cells are B cells.
- Click Classify under Tools in the left panel
- Type B cells for the Name
- Click Save (Figure 29)
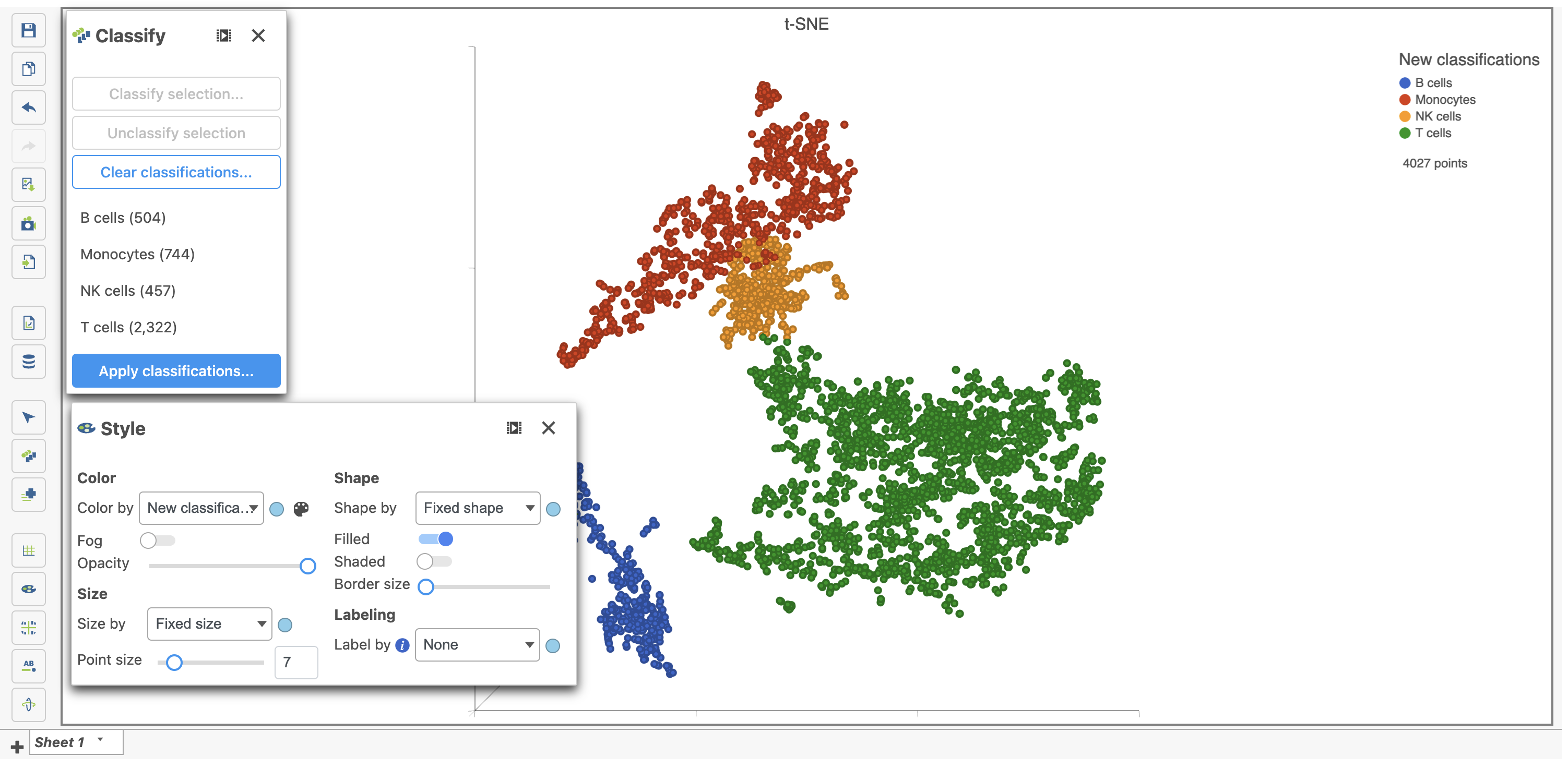
You can edit the name of a classification or delete it. In this project we use the hosted feature lists for "NK cells", "T cells" and "Monocytes" to classify these cell types by coloring the cells in the t-SNE plot and selecting the cells expressing those genes as shown above. See the list management documentation for more information on how to add these lists. The classifications you have made are saved as a working draft so if you close the plot and return to it, the classifications will still be there and can be visualized on the plot as "New classification". However, classifications are not available for downstream tasks until you apply them. Continue classifying the clusters and save the Data viewer session until you are ready to apply the classification to the data project.
- Color by New classifications under Style (Figure 30) while you are still working on the classifications
To use the classifications in downstream tasks and visualizations, you must first apply them.
- Click Apply classifications
- Name the classification (e.g. Classified Cell Types)
- Click Run to confirm
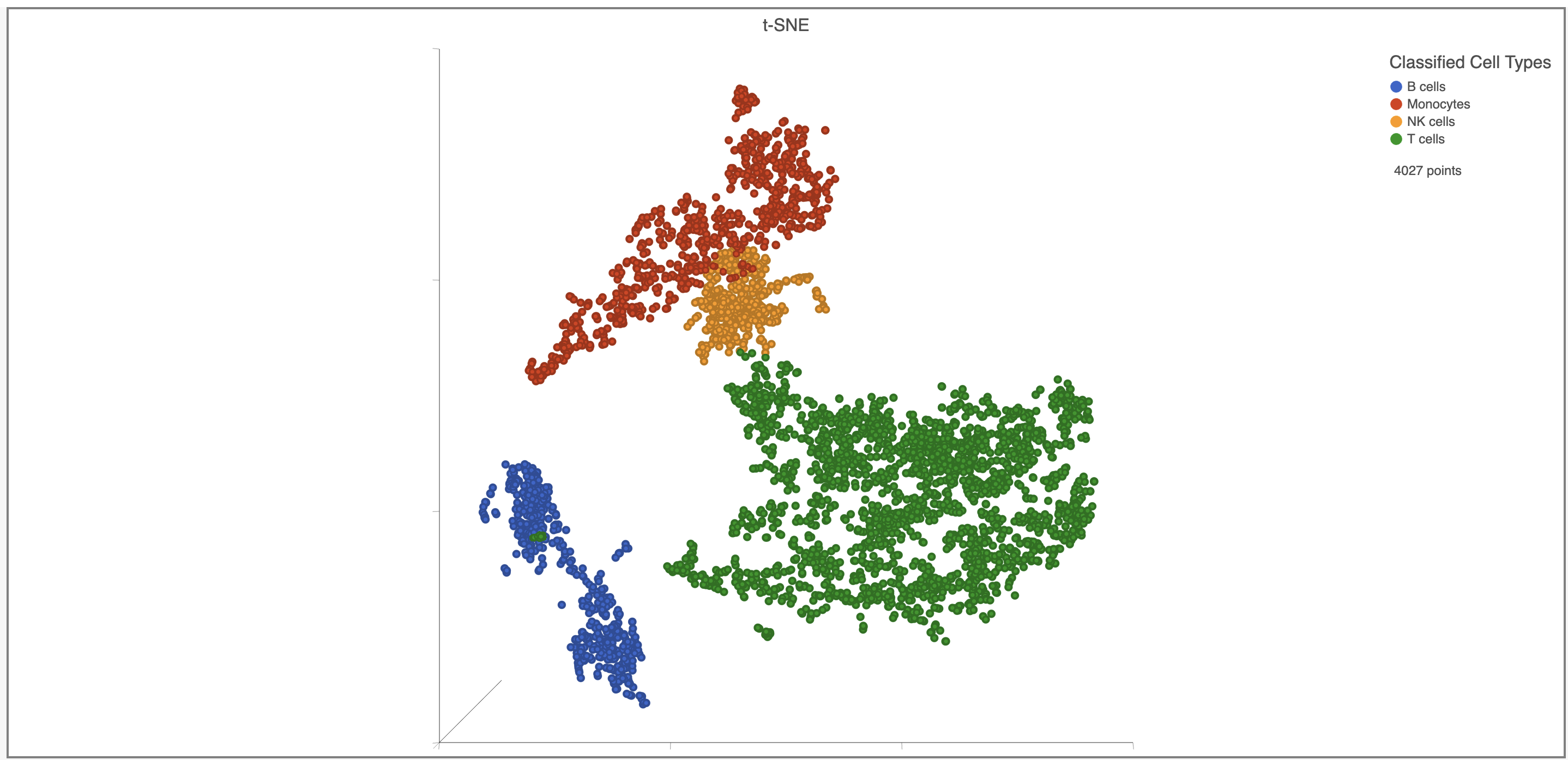
Once you have added a classification to the project, you can color the t-SNE plot by the Classification.
Here, I classified a few additional cell types using a combination of known marker genes and the clustering results then applied the classification (Figure 31).
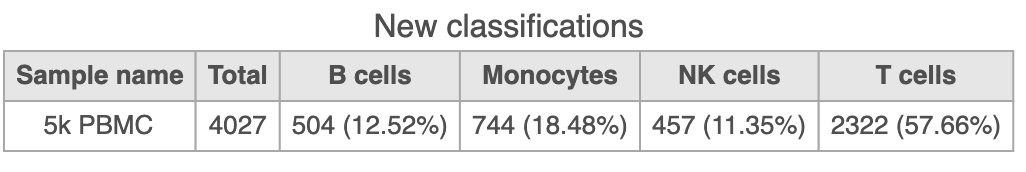
Summarize Classifications with the number and percentage of cells from each sample that belong to each classification using an Attribute table under New plot. This is particularly useful when you are classifying cells from multiple samples.
- Click New plot
- Select Attribute table and the source of data (Figure 32) which in this case is called Classify result
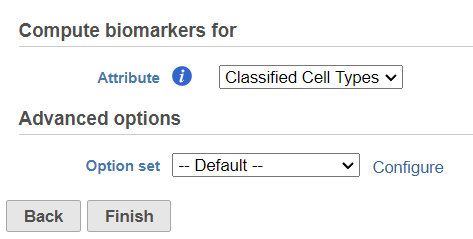
- Click on the Normalized counts" node
- Navigate to the Compute biomarkers task under Statistics in the task menu
- Follow the task dialogue and click Finish (Figure 33)
- Double click the Biomarkers node to view the Biomarkers results
Comparing gene expression between cell types
A common goal in single cell analysis is to identify genes that distinguish a cell type. To do this, you can use the differential analysis tools in Partek Flow. I will show how to use the ANOVA test in Partek Flow, a statistical test shown to be highly effective for differential analysis of single cell RNA-Seq data.
- Click the Normalized counts results node
- Click Statistics in the toolbox
- Click Differential Analysis
- Select ANOVA as the Method to use for differential analysis
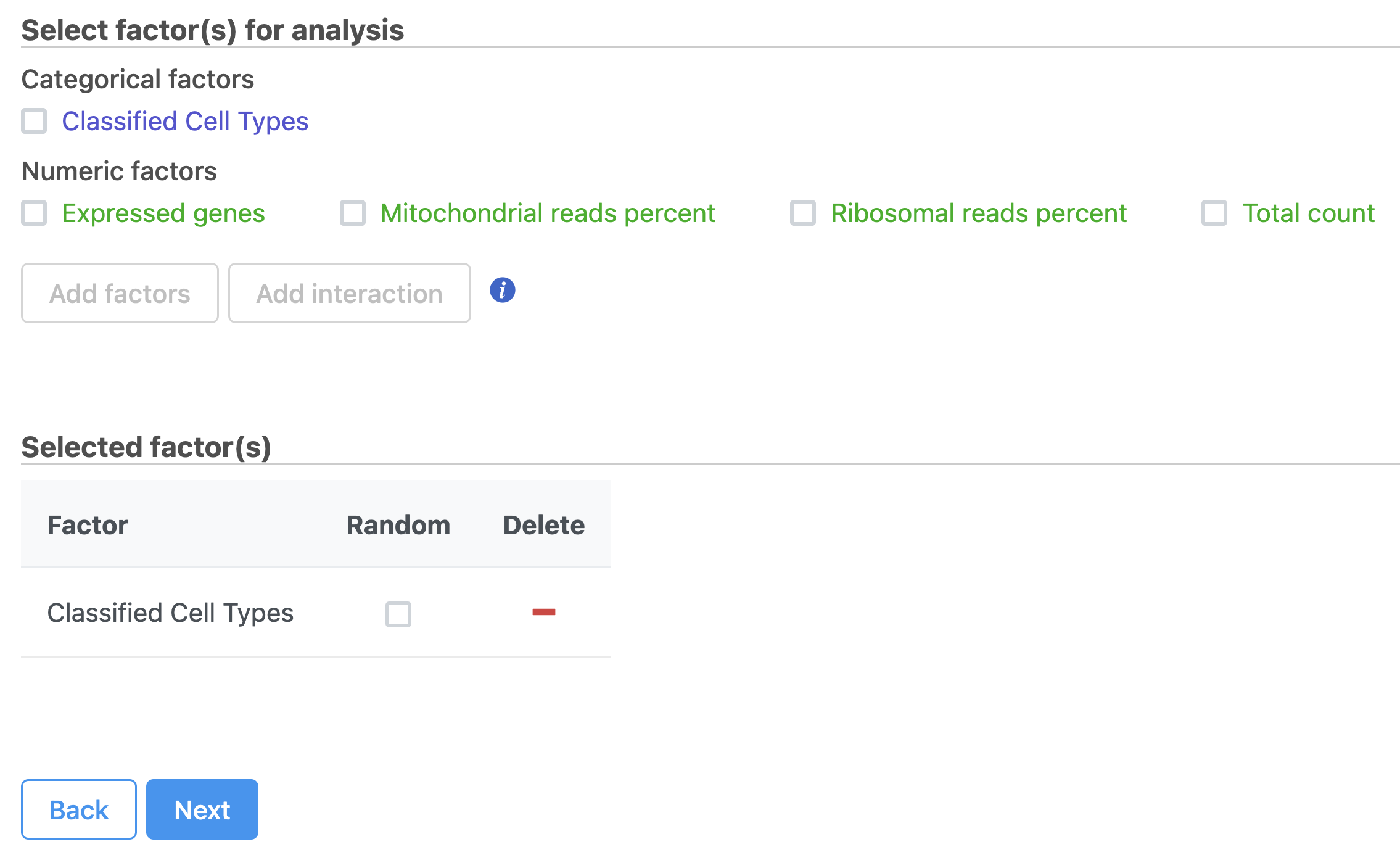
The first page of the configuration dialog asks what attributes you want to include in the statistical test. Here, we only want to consider the Classifications, but in a more complex experiment, you could also include experimental conditions or other sample attributes.
- Click Classified Cell Types
- Click Next (Figure 34)
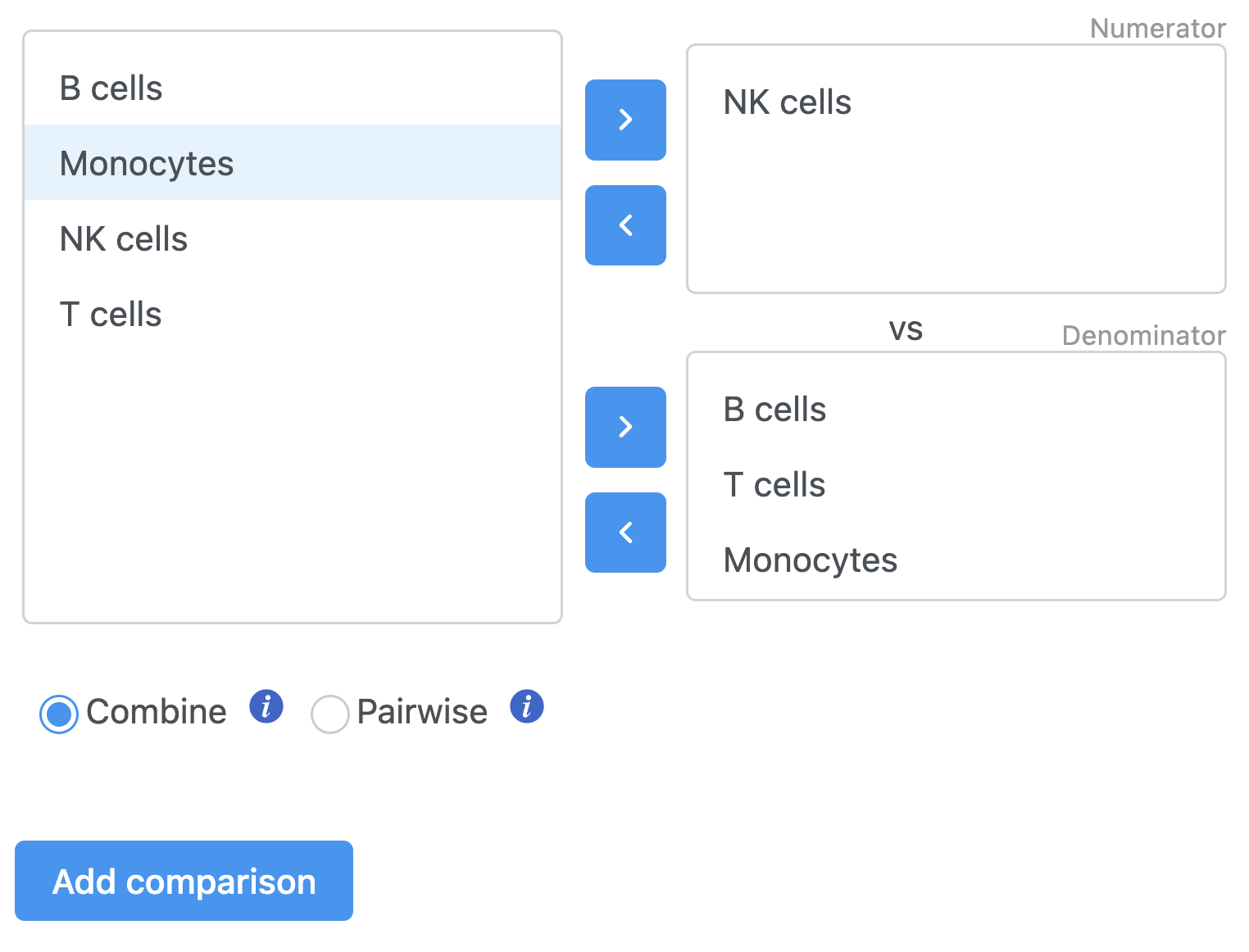
We will make a comparison between NK cells and all the other cell types to identify genes that distinguish NK cells. You can also use this tool to identify genes that differ between two cell types or genes that differ in the same cell type between experimental conditions.
- Drag NK cells to the top panel
The top panel is the numerator for fold-change calculations so the experimental or test groups should be selected in the top panel.
- Click all the other classifications in the bottom panel
The bottom panel is the denominator for fold-change calculations so the control group should be selected in the bottom panel.
- Click Add comparison
This adds the comparison to the statistical test.
- Click Finish to run the ANOVA task (Figure 35)
- Double-click the newly generated data node to open the ANOVA task report
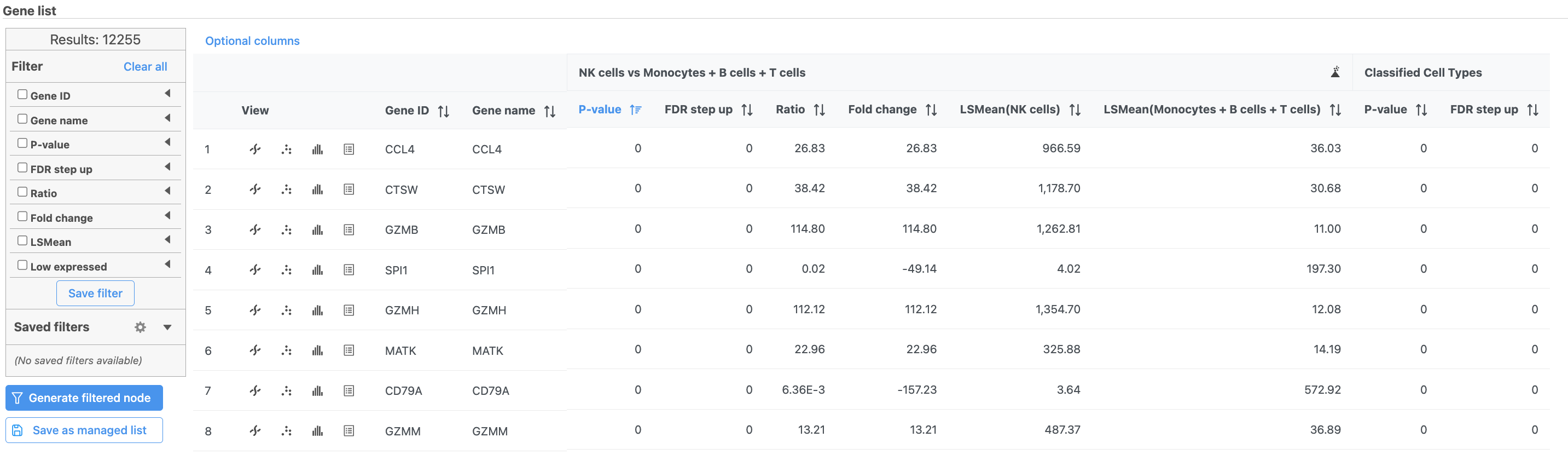
The ANOVA task report lists genes on rows and the results of the statistical test (p-value, fold change, etc.) on columns (Figure 36). For more information, please see our documentation page on the ANOVA task report.
Genes are listed in ascending order by the p-value of the first comparison so the most significant gene is listed first. To view a volcano plot for any comparison, click . To view a violin plot for a gene, click next to the Gene ID.
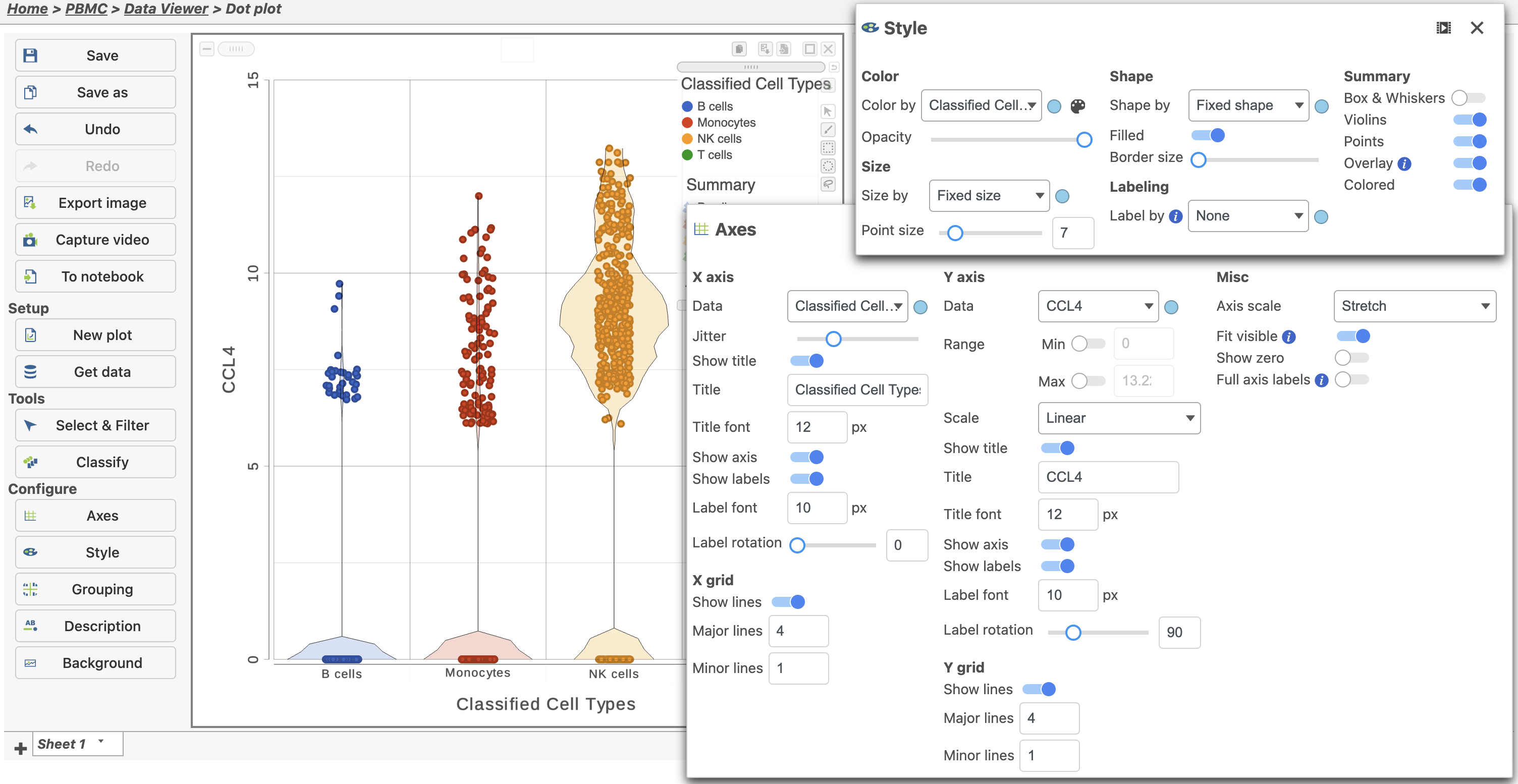
- Click for CCL4
The Feature plot viewer will open showing a dot plot for CCL4 which can be modified to summarize the data in different ways (Figure 37). In the image below, the red boxes highlight the changes that were made to configure the plot. This includes overlaying the violins (density plots with the width corresponding to frequency) on the dot plot represented by the Classified Cell Types.
You can switch the grouping of cells. To do this, show the X axis labels then click and drag the labels to reposition the cell types on the plot.
- Click ANOVA report to return to the table
The table lists all of genes in the data set; using the filter control panel on the left, we can filter to just the genes that are significantly different for the comparison.
- Click FDR step up and click the arrow next to it
- Set to 1e-8
Here, we are using a very stringent cutoff to focus only on genes that are specific to NK cells, but other applications may require a less stringent cutoff.
- Click Fold change and click the arrow next to it
- Set to -2 to 2
The number of genes at the top of the filter control panel updates to indicate how many genes are left after the filters are applied.
- Click to generate a filtered version of the table for downstream analysis
The ANOVA report will close and a new task, the Differential analysis filter, will run and generate a filtered Feature list data node.
For more information about the ANOVA task, please see the Differential Gene Expression - ANOVA section of our user manual.
Generating a heatmap
Once we have filtered to a list of significantly different genes, we can visualize these genes by generating a heatmap.
- Click the Filtered feature list data node produced by the Differential analysis filter
- Click Exploratory analysis in the toolbox
- Click Hierarchical clustering / heatmap
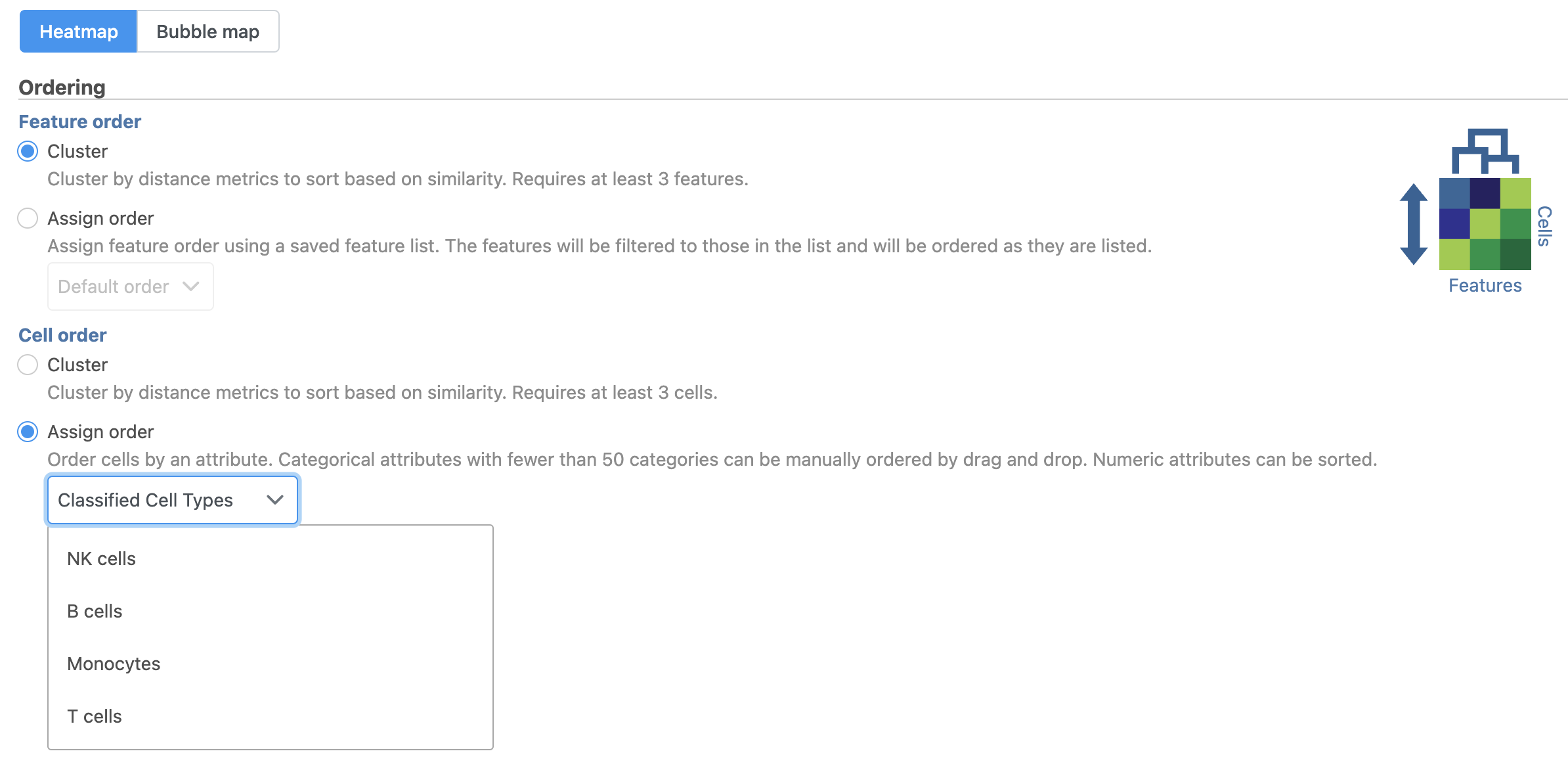
The hierarchical clustering task will generate the heatmap; choose Heatmap as the plot type. You can choose to Cluster features (genes) and cells (samples) under Feature order and Cell order in the Ordering section. You will almost always want to cluster features as this generates the clear blocks of color that make heatmaps comprehensible. For single cell data sets, you may choose to forgo clustering the cells in favor of ordering them by the attribute of interest. Here, we will not filter the cells, but instead order them by their classification.
- Click Assign order under Cell order
You can filter samples using the Filtering section of the configuration dialog. Here, we will not filter out any samples or cells.
- Choose Classification from the Ordering drop-down menu
- Drag NK cells to the top of the Sample order
- Click Finish to run (Figure 38)
- Double-click the Hierarchical cluster task node to open the task report
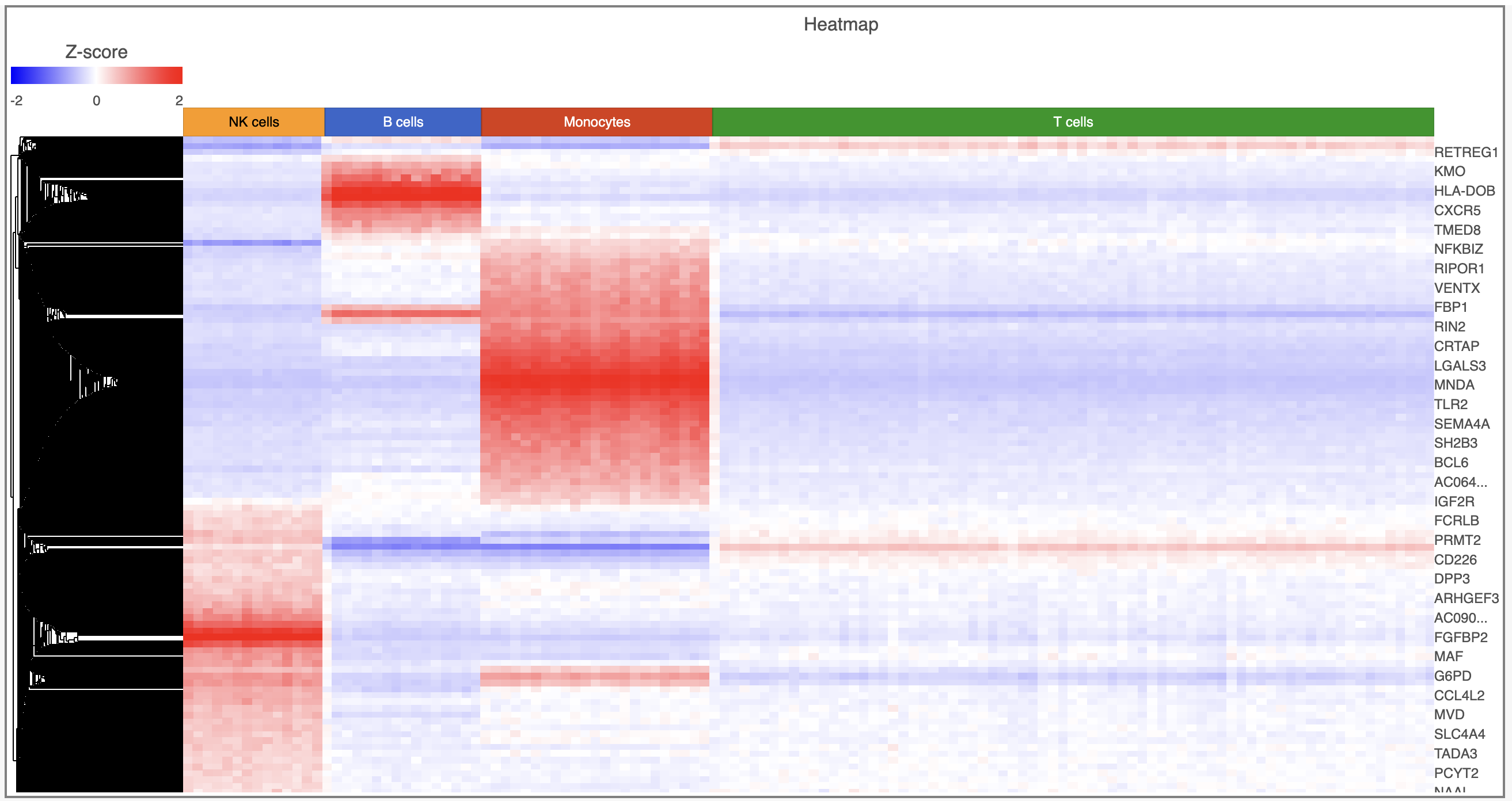
It may initially be hard to distinguish striking differences in the heatmap. This is common in single cell RNA-Seq data because outlier cells will skew the high and low ends. We can adjust the minimum and maximum of the color scheme to improve the appearance of the heatmap.
- Click Heatmap
- Toggle on the Range Min and set to -2
- Toggle on the Range Max and set to 2
Distinct blocks of red and blue are now more pronounced on the plot. Cells are on rows and genes are on columns. Because of the limited number of pixels on the screen, genes are grouped. You can zoom in using the zoom controls or your mouse wheel if you want to view individual gene rows. We can annotate the plot with cell attributes.
- Choose Classified Cell Types from the Annotations drop-down menu
- Change the Annotation font size under Style in the Annotations section
The plot now includes blocks of color along the left edge indicating the classification of the cells. We can transpose the plot to give the cell labels a bit more space.
- Click Transposed under Axes or use the transpose button on the plot to flip the axes
- Toggle off the Row labels under Axes to remove the sample labels
As with any visualization in Partek Flow, the image can be saved as a publication-quality image to your local machine by clicking or sent to a page in the project notebook by clicking . For more information about Hierarchical clustering, please see the Hierarchical Clustering section of the user manual.
Performing enrichment analysis
While a long list of significantly different genes is important information about a cell type, it can be difficult to identify what the biological consequences of these changes might be just by looking at the genes one at a time. Using enrichment analysis, you can identify gene sets and pathways that are over-represented in a list of significant genes, providing clues to the biological meaning of your results.
- Click the Feature list data node produced by the Differential analysis filter
- Click Biological interpretation
- Click Gene set enrichment
We distribute the gene sets from the Gene Ontology Consortium, but Gene set enrichment can work with any custom or public gene set database.
- Choose the latest assembly available from the Gene set drop-down
- Click Finish
- Double-click the Gene set enrichment task node to open the task report
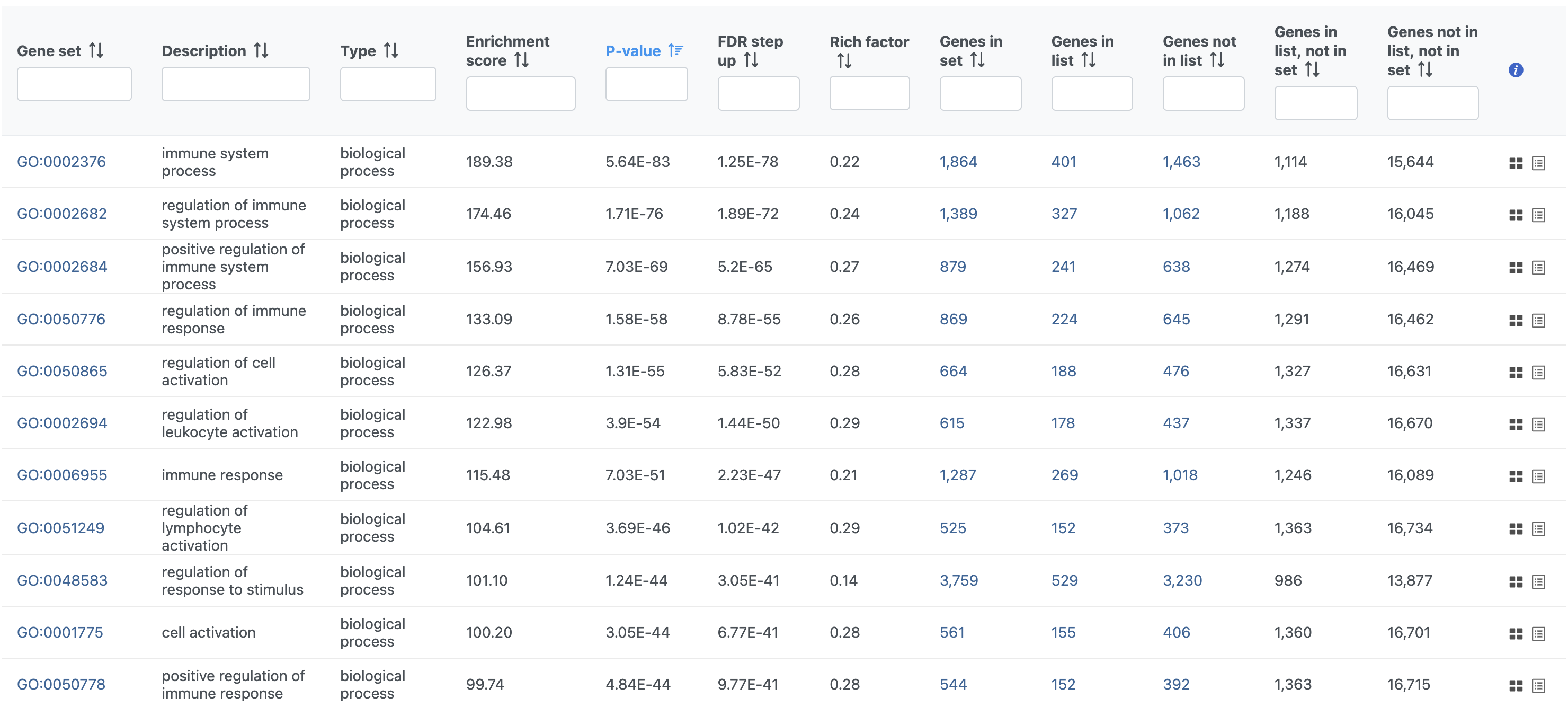
The Gene set enrichment task report lists gene sets on rows with an enrichment score and p-value for each. It also lists how many genes in the gene set were in the input gene list and how many were not (Figure 40). Clicking the Gene set ID links to the geneontology.org page for the gene set.
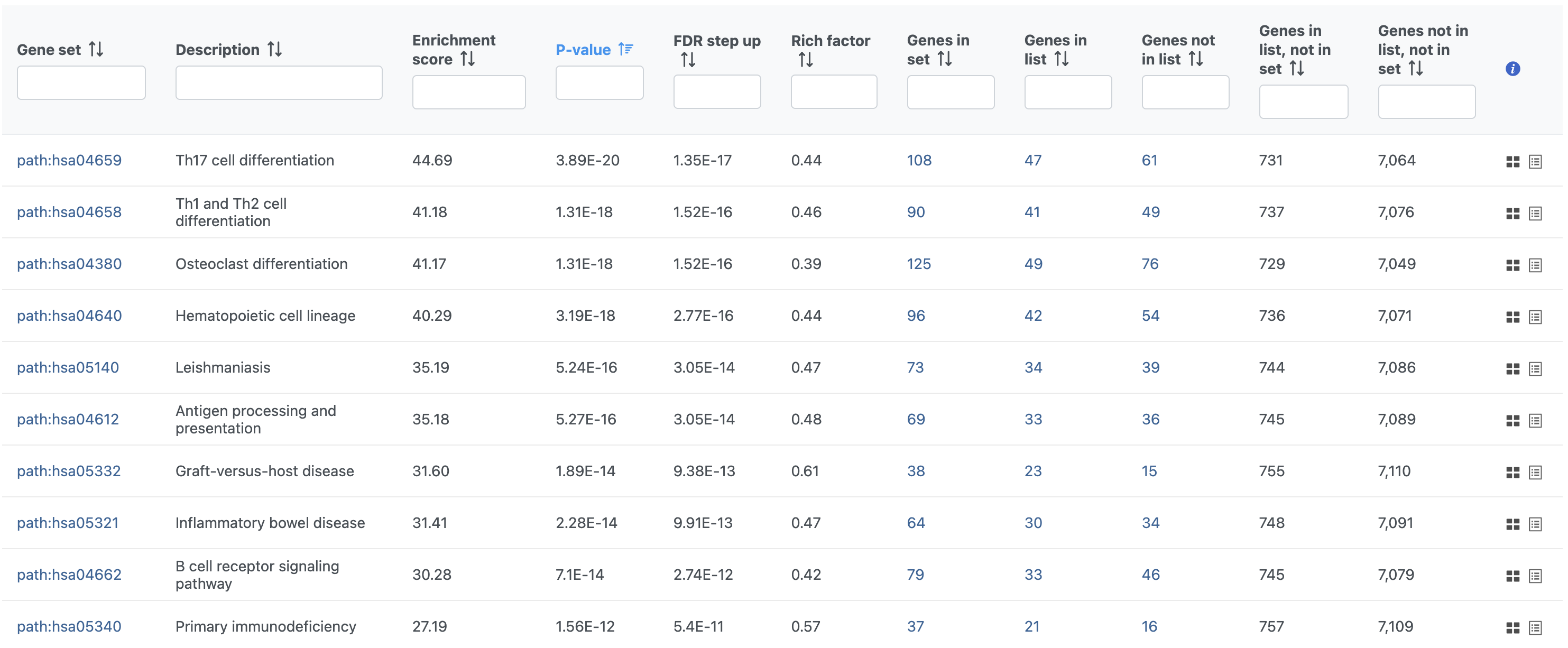
In Partek Flow, you can also check for enrichment of KEGG pathways using the Pathway enrichment task. The task is quite similar to the Gene set enrichment task, but uses KEGG pathways as the gene sets.
The task report is similar to the Gene set enrichment task report with enrichment scores, p-values, and the number of genes in and not in the list (Figure 41).
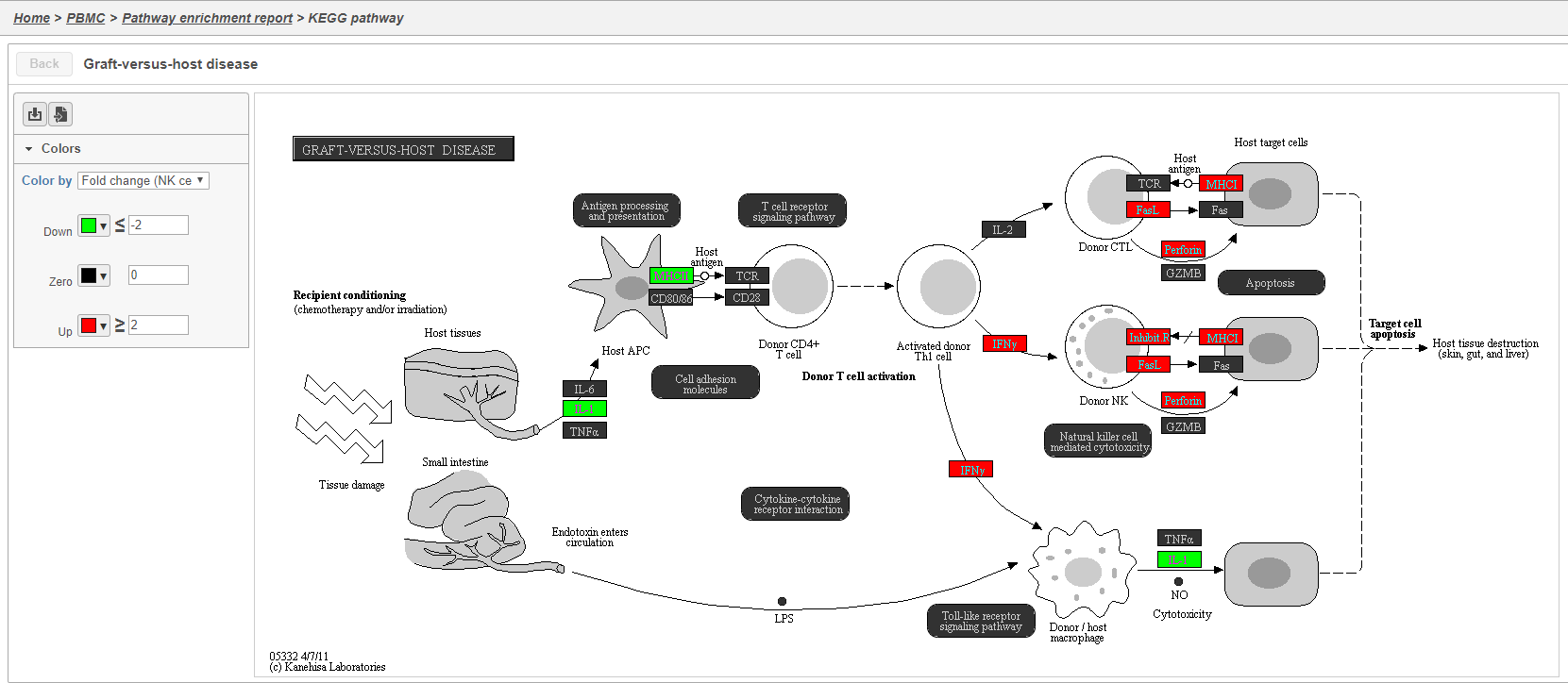
Clicking the KEGG pathway ID in the Pathway enrichment task report opens a KEGG pathway map (Figure 42). The KEGG pathway maps have fold-change and p-value information from the input gene list overlaid on the map, adding a layer of additional information about whether the pathway was upregulated or downregulated in the comparison.
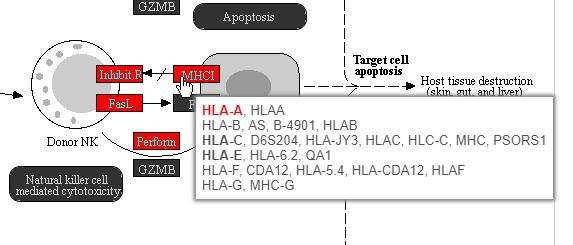
Color are customizable using the control panel on the left and the plot is interactive. Mousing over gene boxes gives the genes accounted for by the box, with genes present in the input list shown in bold, and the coloring gene shown in red (Figure 43).
Clicking a pathway box opens the map of that pathway, providing an easy way to explore related gene networks.
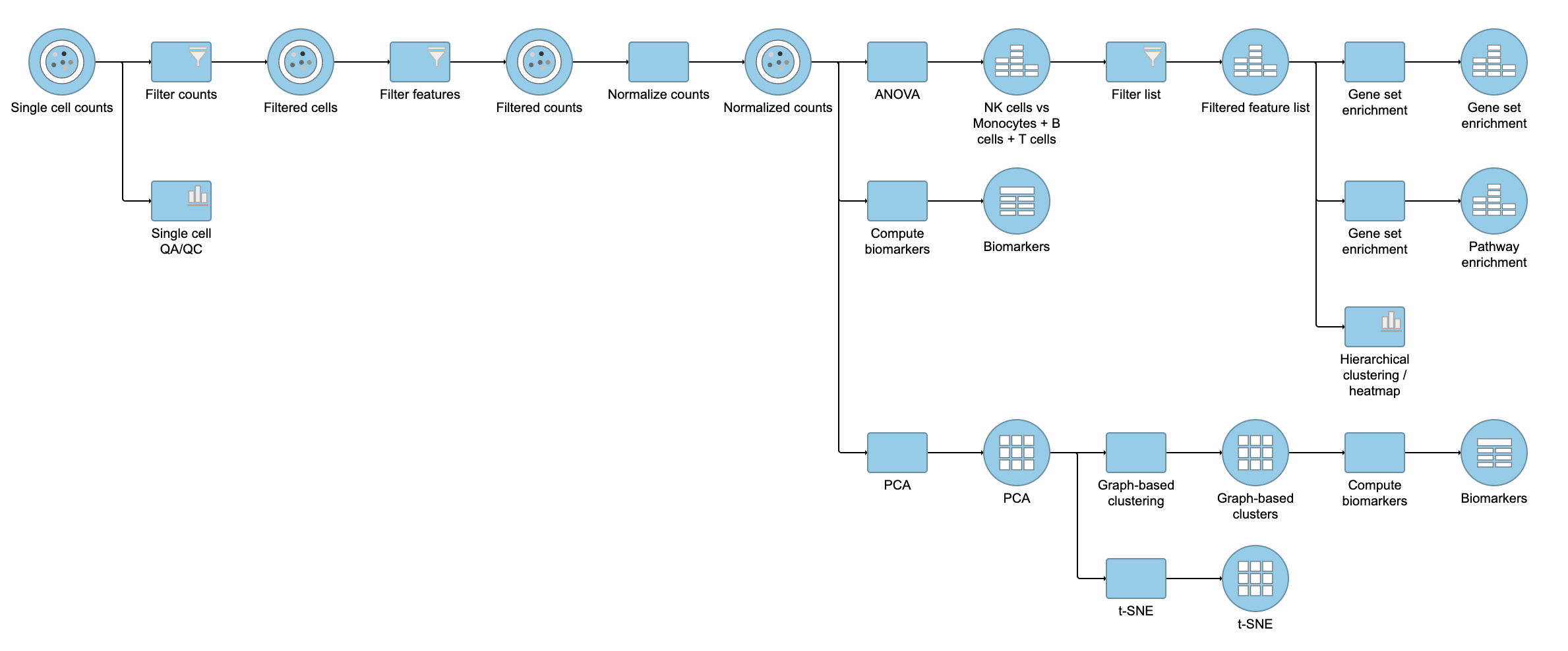
Pipeline
For information about automating steps in this analysis workflow, please see our documentation page on Making a Pipeline.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
29 | rates |