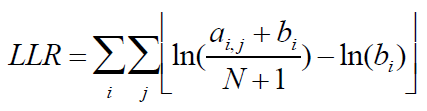Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
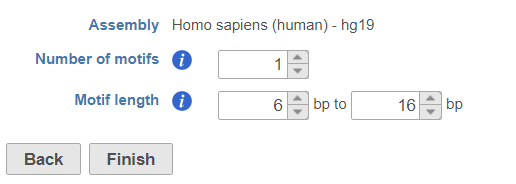
Motif detection tasks allow to extract binding motifs from peak region generated from ChIP-seq data. It allows users to search for know motifs from provided database as well as detect noval motifs. These tasks are available when click on region data node
Detect de novo motifs
Section headings should use level 2 heading, while the content of the section should use paragraph (which is the default). You can choose the style in the first dropdown in toolbar.
Search for know motifs
| Additional assistance |
|---|
|
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
This is done by repeating the below two steps until conveergences:
A. Given the alignment matrix from step B, search for location in the set of regions that score highly compared to the alignment matrix using equation
Search for know motifs
Reference
- Neuwald, A.F., Liu, J.S., & Lawrence, C.E. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Science 1995, 4: 1618-1632.
| Rate Macro | ||
|---|---|---|
|
Overview
Content Tools