Page History
...
- Select Create Marker List from the Analysis section of the Illumina BeadArray Methylation workflow
- Select 1/anova-1way_autosomal_only (ANOVA-1way_autosomal_only) as the source spreadsheet from the left-hand panel
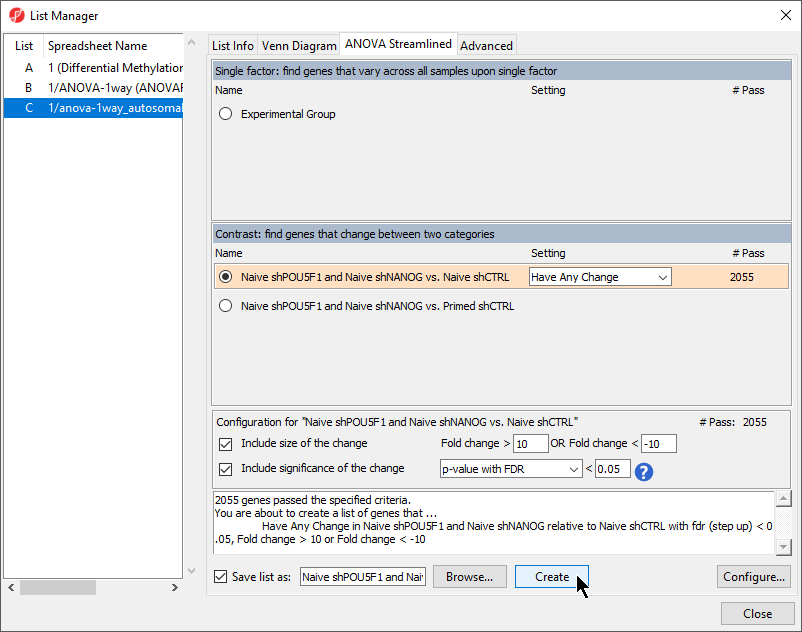
- Select Naive shPOU5F1 and Naive shNANOG and Primed shCTRL vs. Naive shCTRL from the Contrast: find genes that change between two categories section of the ANOVA Streamlined tab of the List Manager dialog
- Set Fold change > to 10 3
- Set OR Fold change < to -103
- Leave the rest of the option set to defaults (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Create
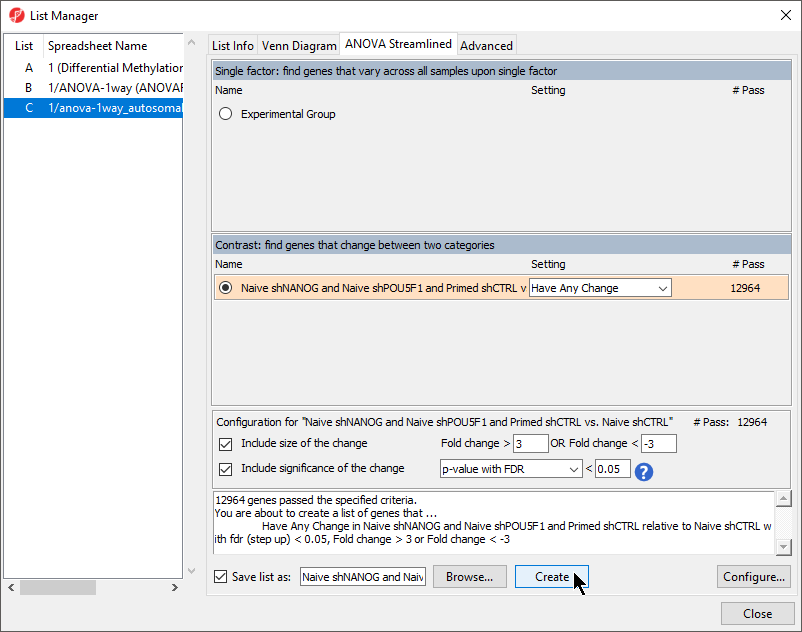
- Create another marker list with the same criteria for the contrast Naive shPOU5F1 and Naive shNANOG vs. Primed shCTRL
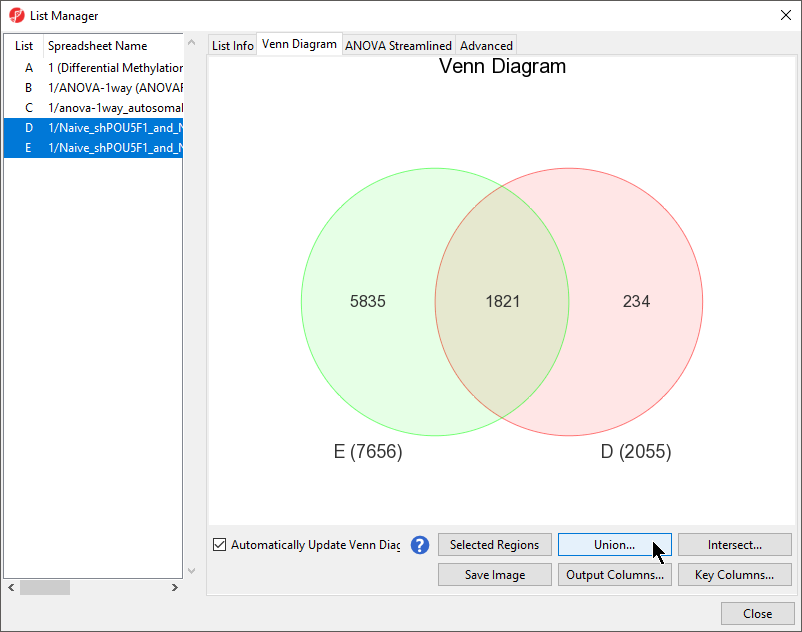
- Select the Ven Diagram tab on the List Manager dialog
- Select both D and E from as the source spreadsheets from the left-hand panel list by holding Ctrl and selecting both D and E
- Select Union... to create a list with probes from both spreadsheets (Figure 2)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
- Select Close to exit the List Manager dialog
- Select () from the quick action bar to save the newly created spreadsheet
- Specify a name for the spreadsheet, we chose CpG Loci of Interest, using the Save File dialog
- Select Save to save the spreadsheet
You may want to save the project before proceeding to the next section of the tutorial.
...
Overview
Content Tools