Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
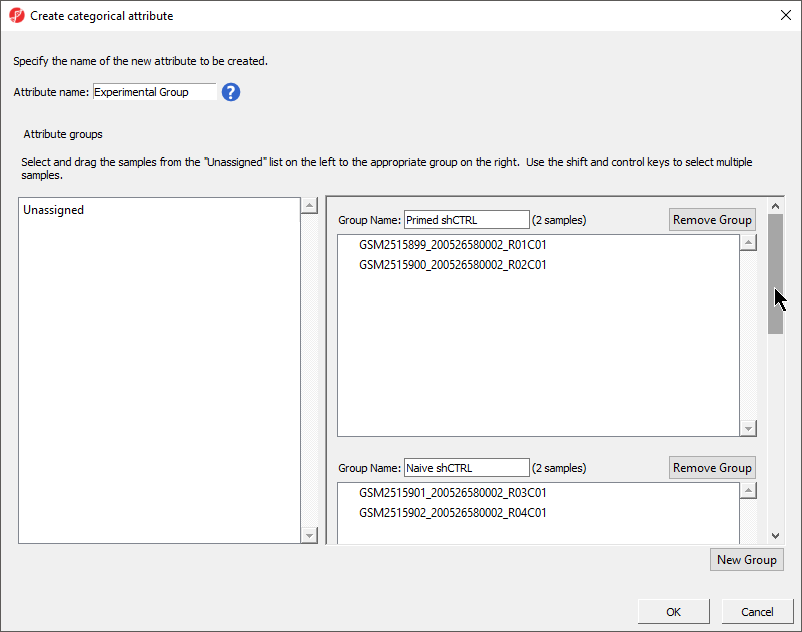
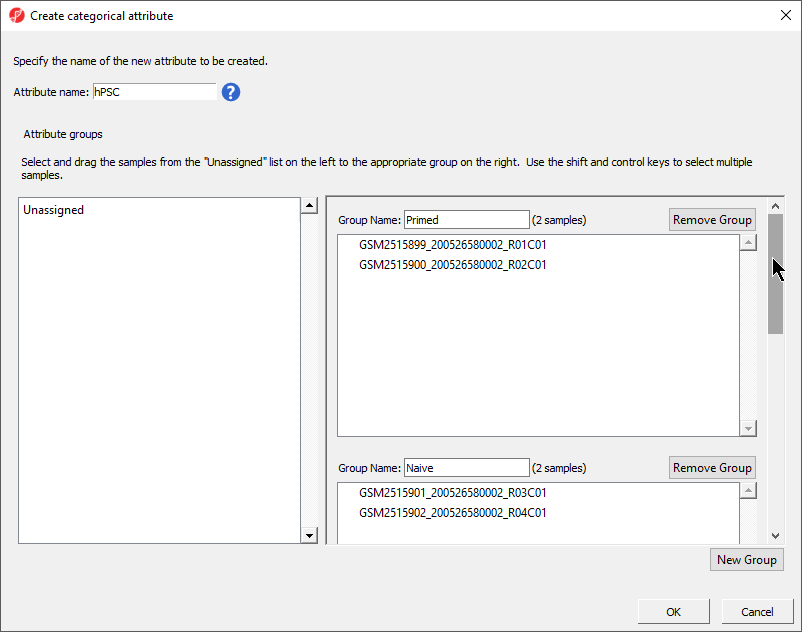
- Select New Group twice to add two additional groups
- Set Set Attribute name: to hPSC State
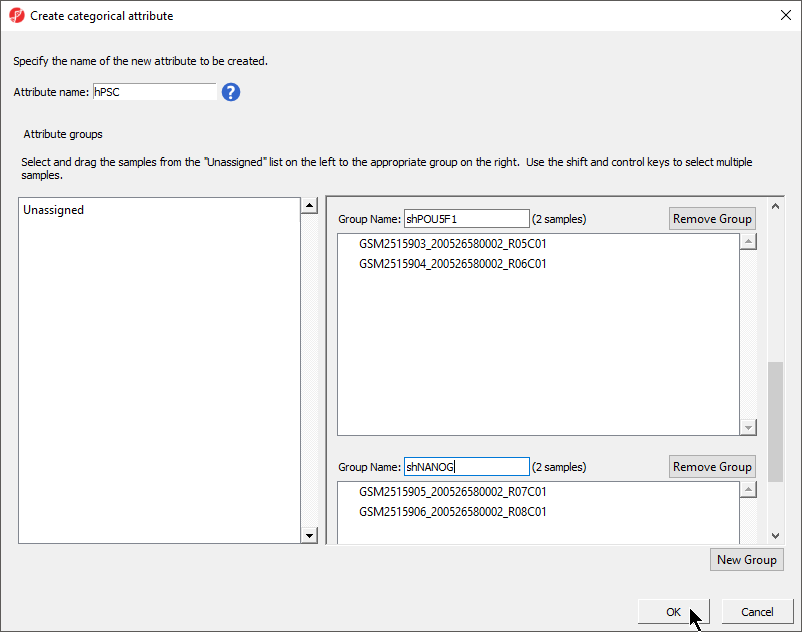
- Rename the four groups Primed, Naive, shPOU5F1, and shNANOGPrimed and Naive
- Drag and drop the samples from the Unassigned list to their groups as listed in the table below
| Sample ID | Group Name |
|---|---|
| GSM2515899_200526580002_R01C01 | Primed |
| GSM2515900_200526580002_R02C01 | Primed |
| GSM2515901_200526580002_R03C01 | Naive |
| GSM2515902_200526580002_R04C01 | Naive |
| GSM2515903_200526580002_R05C01 | shPOU5F1Naive |
| GSM2515904_200526580002_R06C01 | shPOU5F1Naive |
| GSM2515905_200526580002_R07C01 | shNANOGNaive |
| GSM2515906_200526580002_R08C01 | shNANOGNaive |
There should now be four two groups with two samples in each group (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| ||||
- Select OK
- Select Yes from the Add another categorical attribute dialog
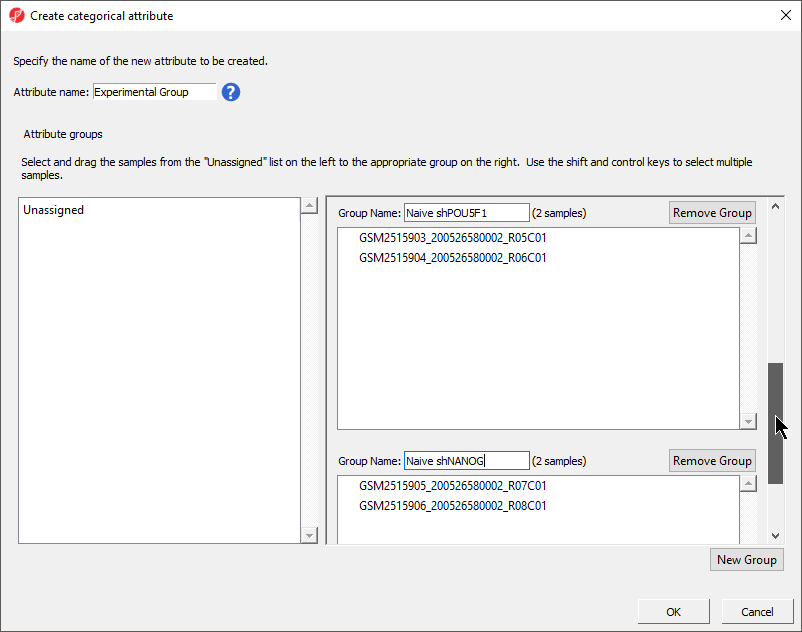
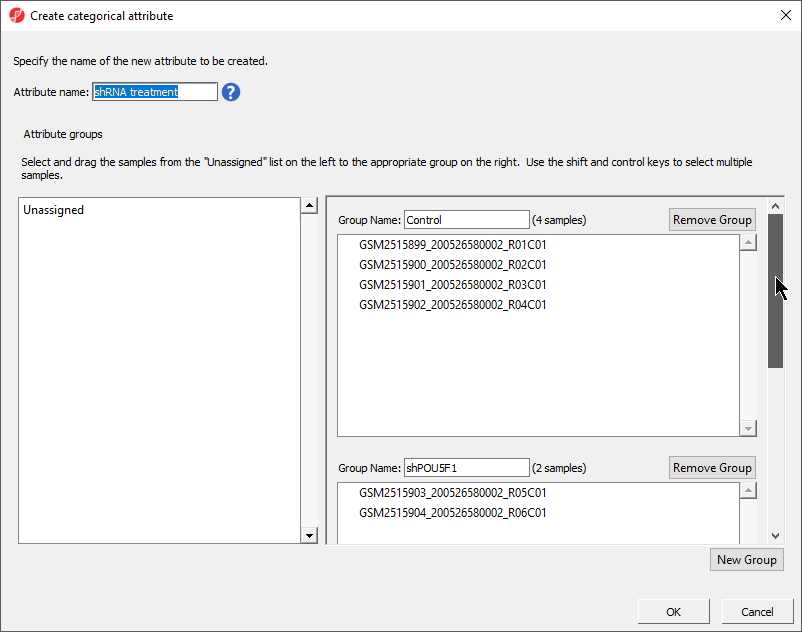
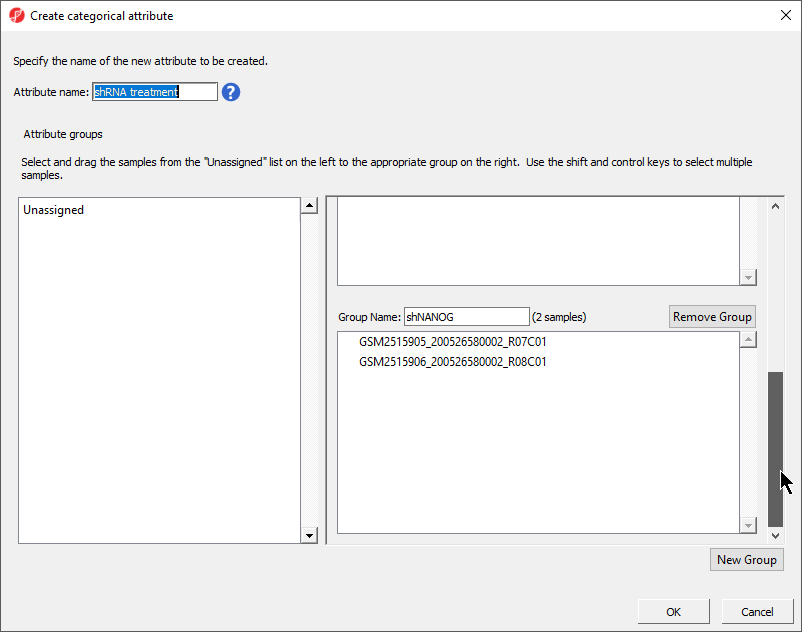
- Set Attribute name: to shRNA treatment
- Select New Group to add an additional group
- Rename the three groups Control, shPOU5F1, and shNANOG
- Drag and drop the samples from the Unassigned list to their groups as listed in the table below
| Sample ID | Group Name |
|---|---|
GSM2515899_200526580002_R01C01 | Control |
GSM2515900_200526580002_R02C01 | Control |
GSM2515901_200526580002_R03C01 | Control |
GSM2515902_200526580002_R04C01 | Control |
GSM2515903_200526580002_R05C01 | shPOU5F1 |
GSM2515904_200526580002_R06C01 | shPOU5F1 |
GSM2515905_200526580002_R07C01 | shNANOG |
GSM2515906_200526580002_R08C01 | shNANOG |
There should now be three groups with samples in each group (Figure 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select OK
- Select No from the Add another categorical attribute dialog
- Select Yes to save the spreadsheet
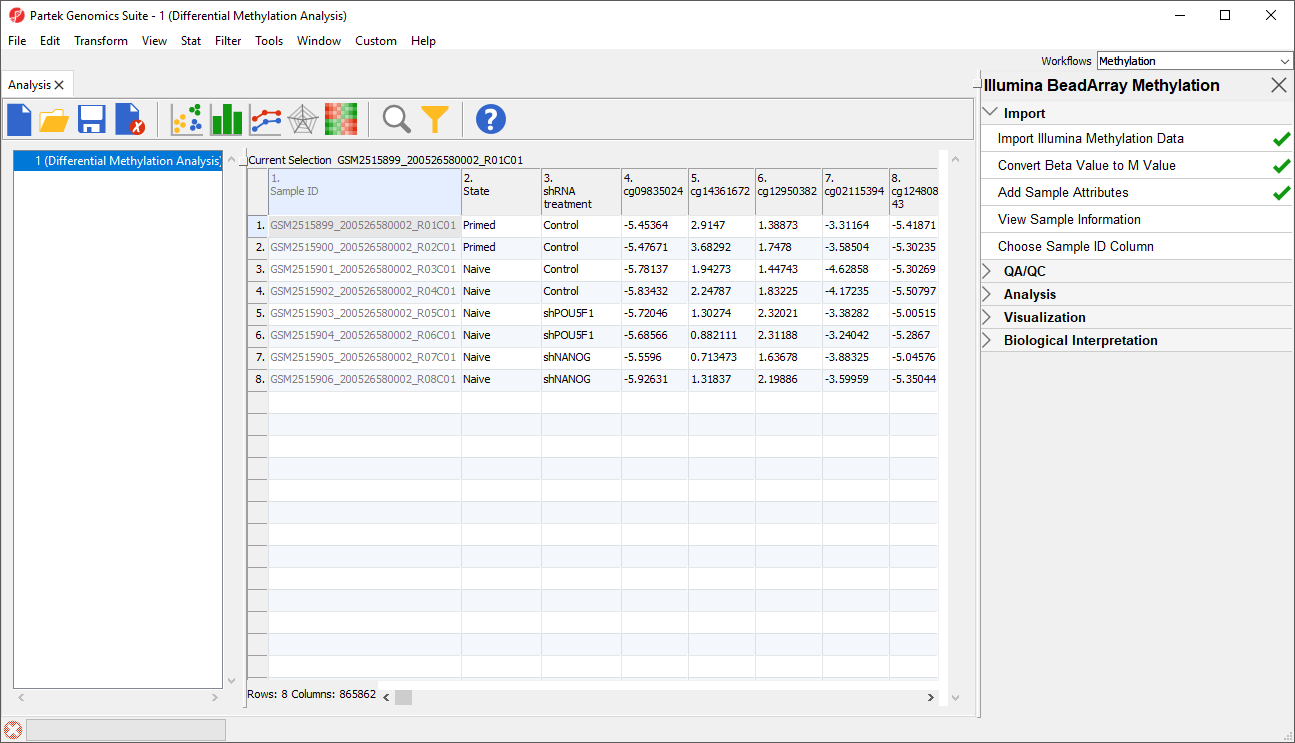
A Two new column as been added to spreadsheet 1 (Differential Methylation Analysis) with the experimental group state and shRNA treatment of each sample (Figure 56).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Page Turner | ||
|---|---|---|
|
...
Overview
Content Tools