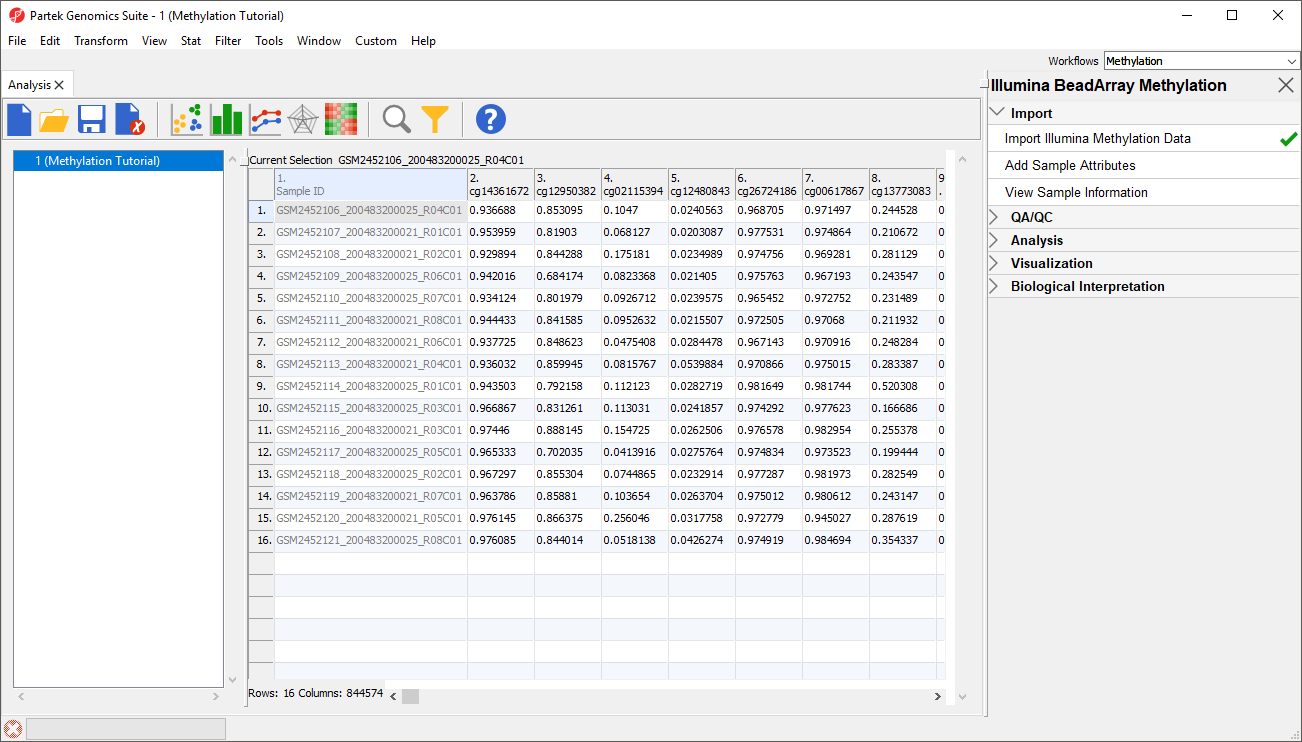
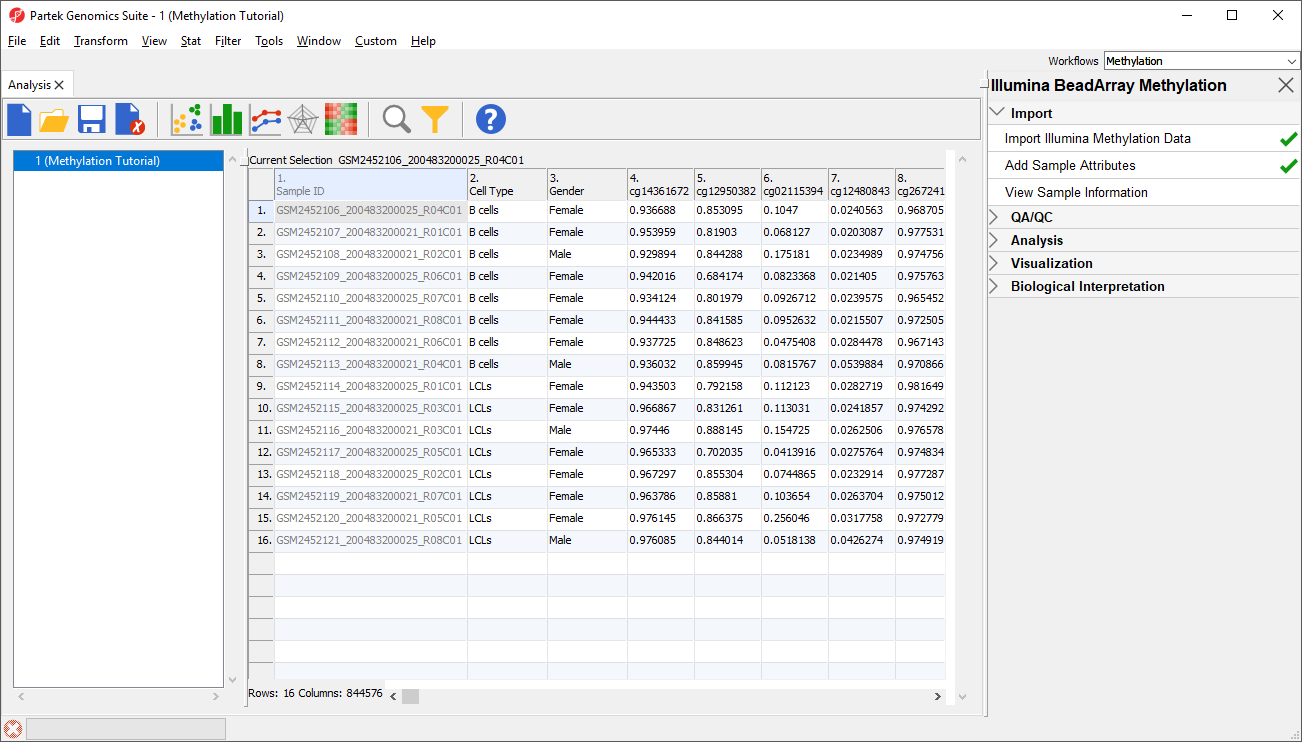
Each row of the spreadsheet (Figure 1) corresponds to a single sample. The first column is the names of the .idat files and the remaining columns are the array probes. The table values are β-values, which correspond to the percentage methylation at each site. A β-value is calculated as the ratio of methylated probe intensity over the overall intensity at each site (the overall intensity is the sum of methylated and unmethylated probe intensities).
Before we can perform any analysis, the study samples need to be organized into their experimental groups.
- Select Add Sample Attributes from the Import section of the Illumina BeadArray Methylation workflow
- Select Add a Categorical Attribute from the Add Sample Attributes dialog (Figure 2)
- Select OK
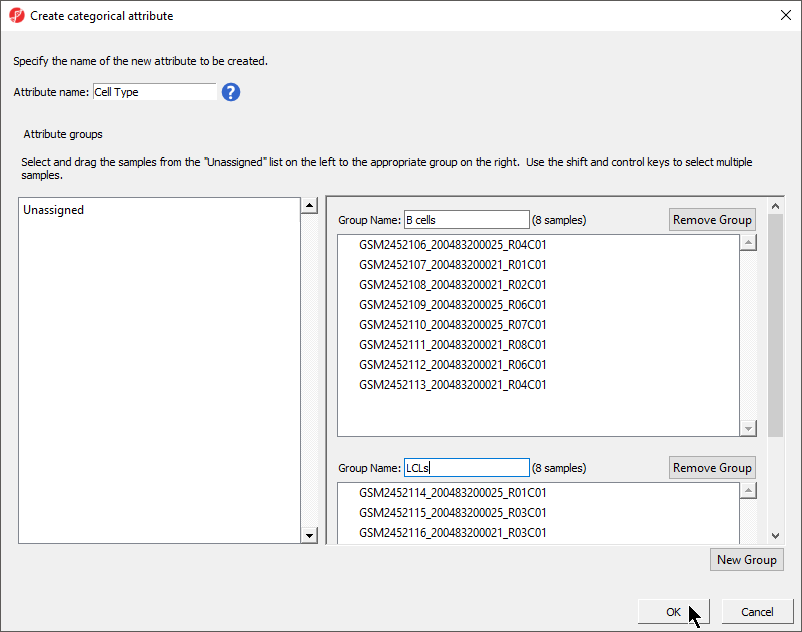
The Create categorical attribute dialog allows us to create groups for a categorical attribute. By default, two groups are created, but additional groups can be added.
- Set Attribute name: to Cell Type
- Rename the groups B cells and LCLs
- Drag and drop the samples from the Unassigned list to their groups as listed in the table below
| Sample ID | Cell Type |
|---|---|
| GSM2452106_200483200025_R04C01 | B cells |
| GSM2452107_200483200021_R01C01 | B cells |
| GSM2452108_200483200021_R02C01 | B cells |
| GSM2452109_200483200025_R06C01 | B cells |
| GSM2452110_200483200025_R07C01 | B cells |
| GSM2452111_200483200021_R08C01 | B cells |
| GSM2452112_200483200021_R06C01 | B cells |
| GSM2452113_200483200021_R04C01 | B cells |
| GSM2452114_200483200025_R01C01 | LCLs |
| GSM2452115_200483200025_R03C01 | LCLs |
| GSM2452116_200483200021_R03C01 | LCLs |
| GSM2452117_200483200025_R05C01 | LCLs |
| GSM2452118_200483200025_R02C01 | LCLs |
| GSM2452119_200483200021_R07C01 | LCLs |
| GSM2452120_200483200021_R05C01 | LCLs |
| GSM2452121_200483200025_R08C01 | LCLs |
There should now be two groups with eight samples in each group (Figure 3).
- Select OK
- Select Yes from the Add another categorical attribute dialog
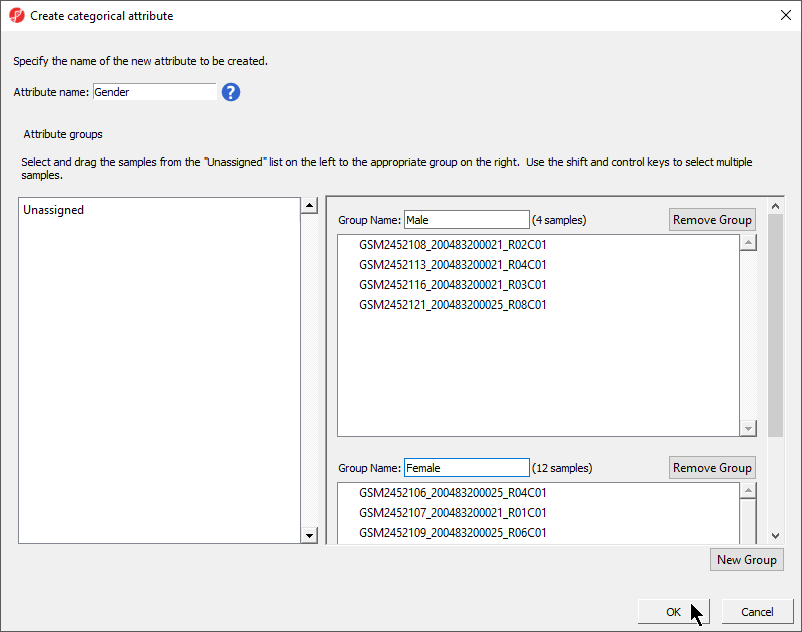
- Set Attribute name: to Gender
- Rename the groups Male and Female
- Drag and drop the samples from the Unassigned list to their groups as listed in the table below
| Sample ID | Gender |
|---|---|
| GSM2452106_200483200025_R04C01 | Female |
| GSM2452107_200483200021_R01C01 | Female |
| GSM2452108_200483200021_R02C01 | Male |
| GSM2452109_200483200025_R06C01 | Female |
| GSM2452110_200483200025_R07C01 | Female |
| GSM2452111_200483200021_R08C01 | Female |
| GSM2452112_200483200021_R06C01 | Female |
| GSM2452113_200483200021_R04C01 | Male |
| GSM2452114_200483200025_R01C01 | Female |
| GSM2452115_200483200025_R03C01 | Female |
| GSM2452116_200483200021_R03C01 | Male |
| GSM2452117_200483200025_R05C01 | Female |
| GSM2452118_200483200025_R02C01 | Female |
| GSM2452119_200483200021_R07C01 | Female |
| GSM2452120_200483200021_R05C01 | Female |
| GSM2452121_200483200025_R08C01 | Male |
There should now be two groups with four samples in Male and twelve samples in Female (Figure 4).
- Select OK
- Select No from the Add another categorical attribute dialog
- Select Yes to save the spreadsheet
Two new columns have been added to spreadsheet 1 (Methylation) with the cell type and gender of each sample (Figure 5).
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
35 | rates |