Page History
...
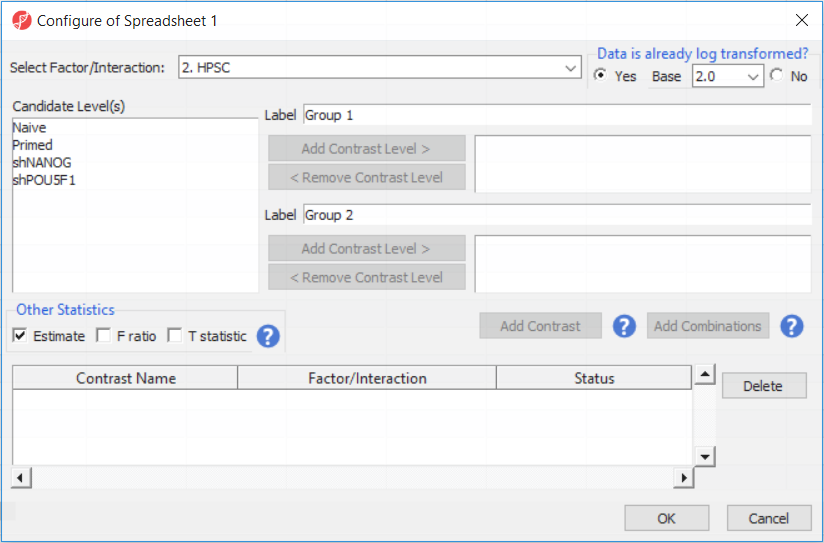
Depending on the data in the top level (i.e. parent) spreadsheet you may want to manually specify whether the data is already log transformed: you should select Yes for M-values (but otherwise No for β-values) (Figure 2). By default, Partek Genomics Suite will calculate fold-change value for each contrast . In addition(since M-values are in log2 space, the resulting fold-change is actually a difference in methylation levels). On the other hand, to include the difference in methylation levels at each CpG site in the output (i.e. ΔM or Δβquite common for β-values), check the Estimate box in the Other Statistics section of the dialog (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Overview
Content Tools