Page History
...
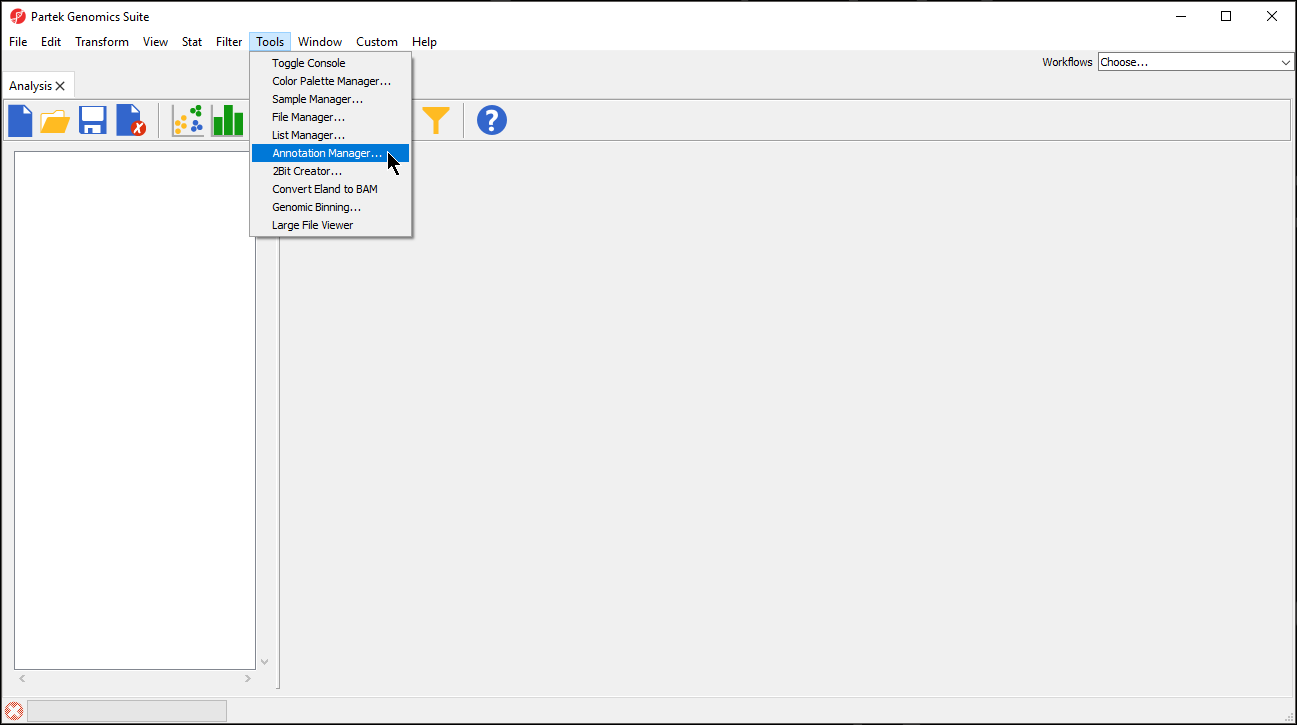
- Select Tools from the main toolbar
- Select Annotation Manager... (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
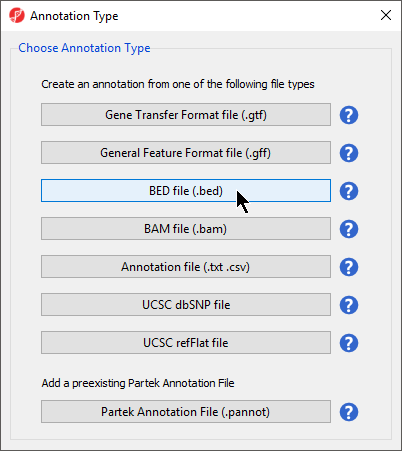
- Select BED file (.bed) for Annotation Type (Figure 23)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
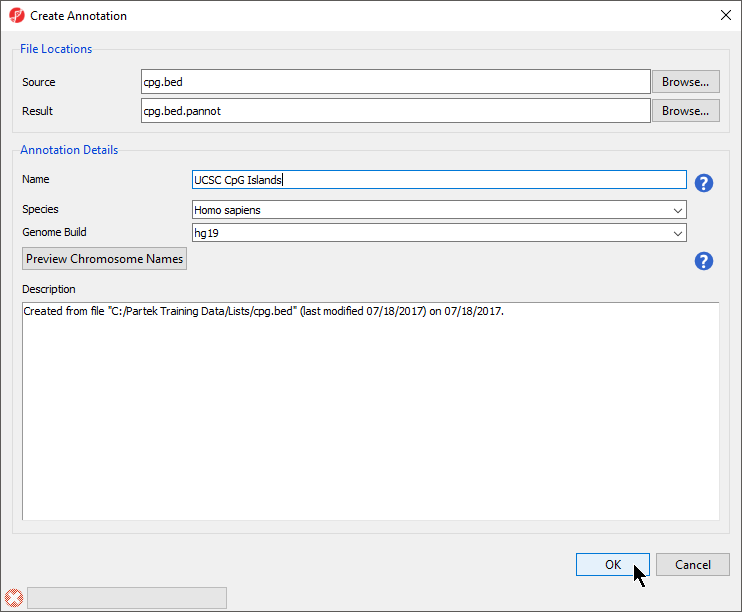
- Select Browse... under Source to specify the BED file; a default new file name and destination will populate Result, but this can be changed
- You can specify the name and save location of the new annotation file under Result; we typically choose the Microarray Libraries folder
- Specify the Name of the annotation database file
- Select the correct Species and Genome Build for the annotation file from the drop-down menus (Figure 34)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
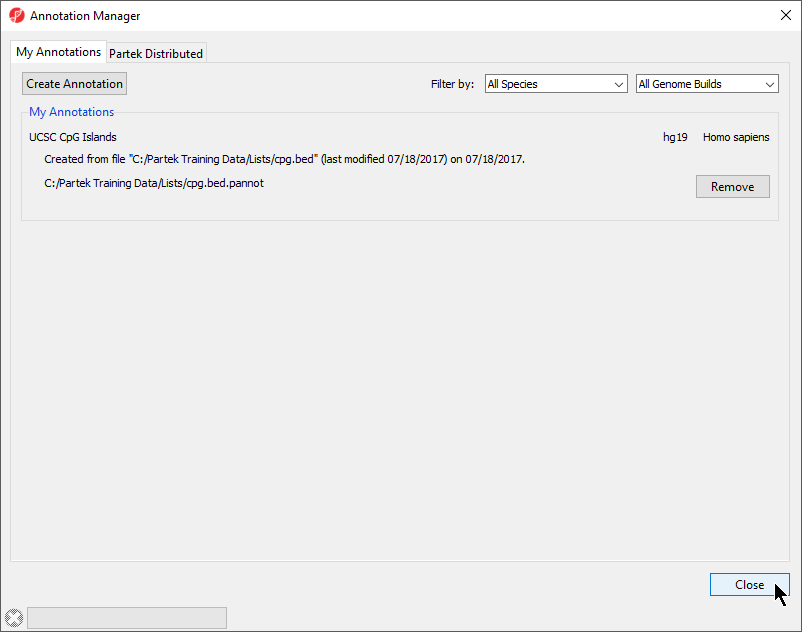
The Annotation Manager will display the new annotation in the My Annotations tab (Figure 45)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Visualizing a BED file as an annotation track in the genome browser
...
- Right-click a row on any spreadsheet that has genomic features on rows (gene lists, ANOVA results, SNP detection)
- Select either Browse to Row or Browse to Location to invoke the Genome Browser tab
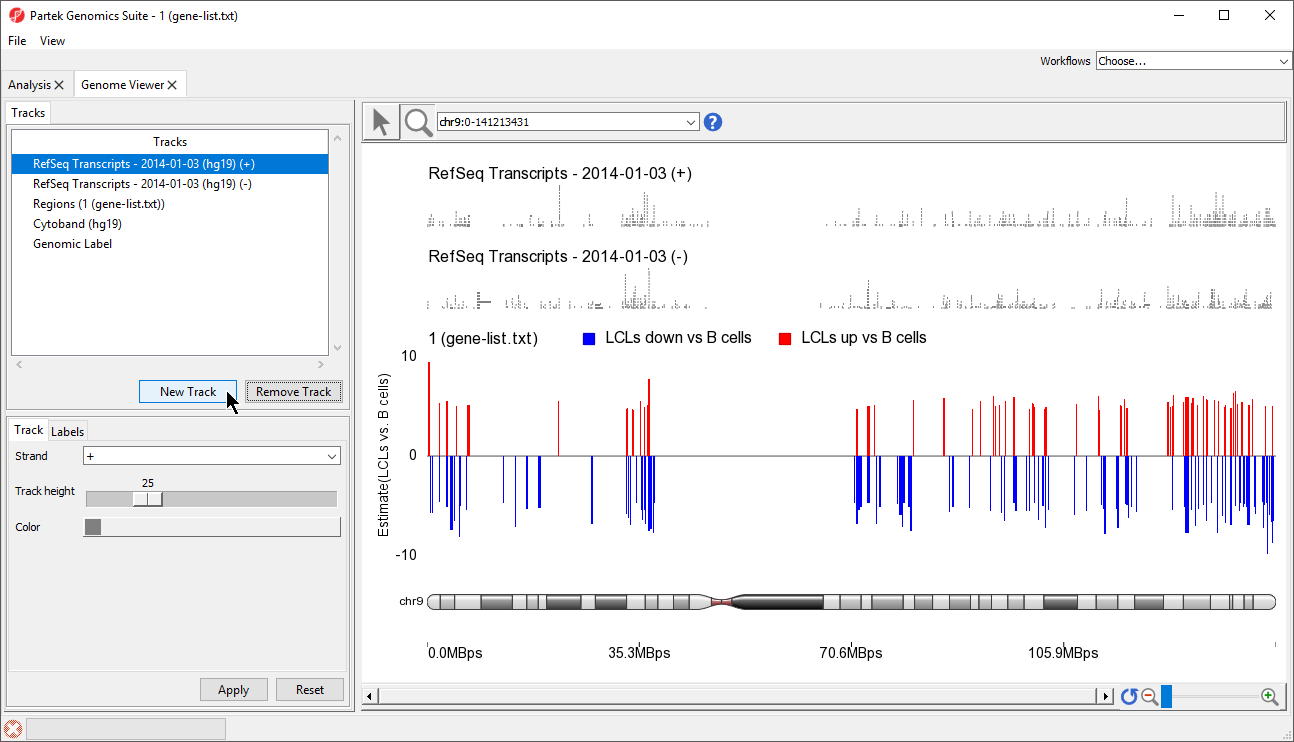
- Select New Track from the Tracks panel of the Genome Browser (Figure 56)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
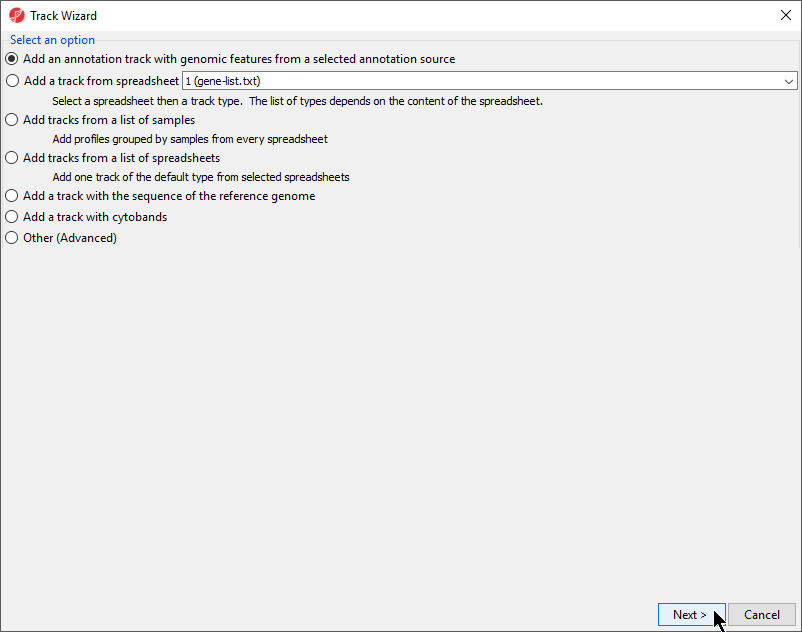
- Select Add an annotation track with genomic features from a selected annotation source from the Track Wizard dialog (Figure 67)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Next >
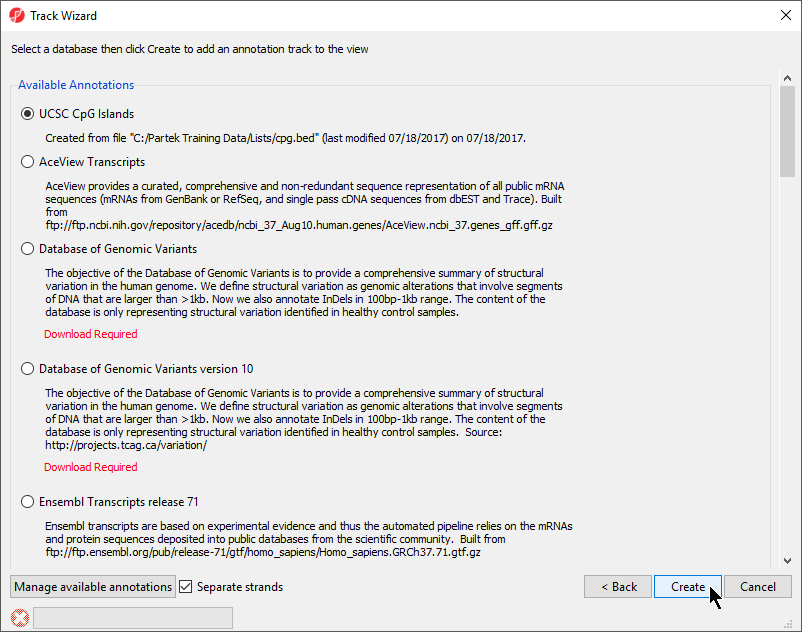
- Choose the annotation file you created; here we have selected UCSC CpG Islands (Figure 78)
- If your annotation file does not contain strand information for each region, deselect Separate Strands; here we have deselected it
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
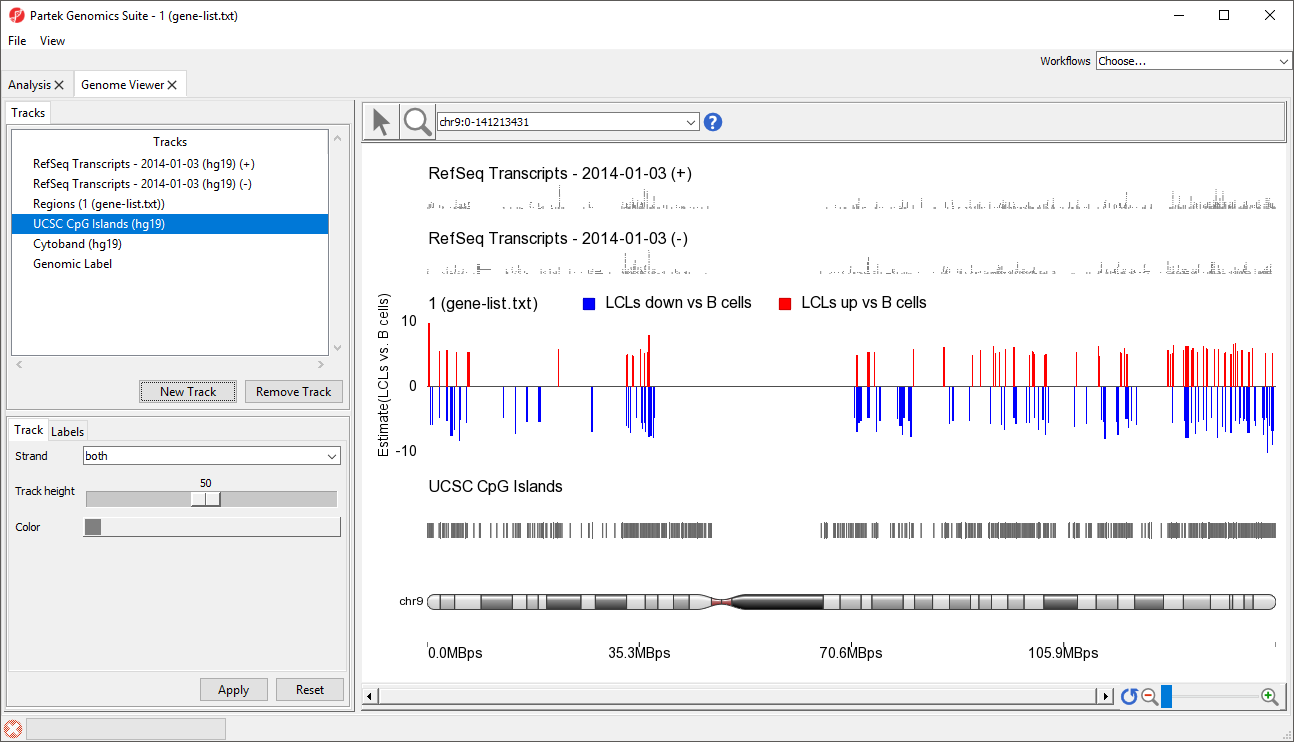
A new track will be created from the annotation file (Figure 89). If Separate Strands had been selected, there would be two tracks, one for each strand, like we see for the RefSeq Transcripts - 2014-01-03 (+) and (-) tracks (Figure 8).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Page Turner | ||
|---|---|---|
|
...
Overview
Content Tools