A BED (Browser Extensible Data) file is a special case of a region list: it is a tab-delimited text file and the first three columns of BED files contain the chromosome, start, and stop locations. To import a bed file to be used as a data region list, follow the import instructions for region lists. A BED File might also be visualized as an annotation file containing regions in the Genome Browser.
BED files do not contain individual sequences nor do the regions have names. For example, the UCSC Genome Browser has an annotation BED file for CpG islands. You might like to view this information in the context of a methylation microarray data set. Before you can visualize a BED file in the chromosome viewer, you must create a Partek annotation file from the BED file.
|
|
|
|
Preview Chromosome Names would be used if the original file had chromosome names that did not match the genome build that had required modification. For our example, this is unnecessary.
The Annotation Manager will display the new annotation in the My Annotations tab (Figure 5)
|
In order to use a BED file as an Annotation track in the Genome Browser, first create the annotation file as described above, being careful to specify the correct species and genome build.
 |
|
|
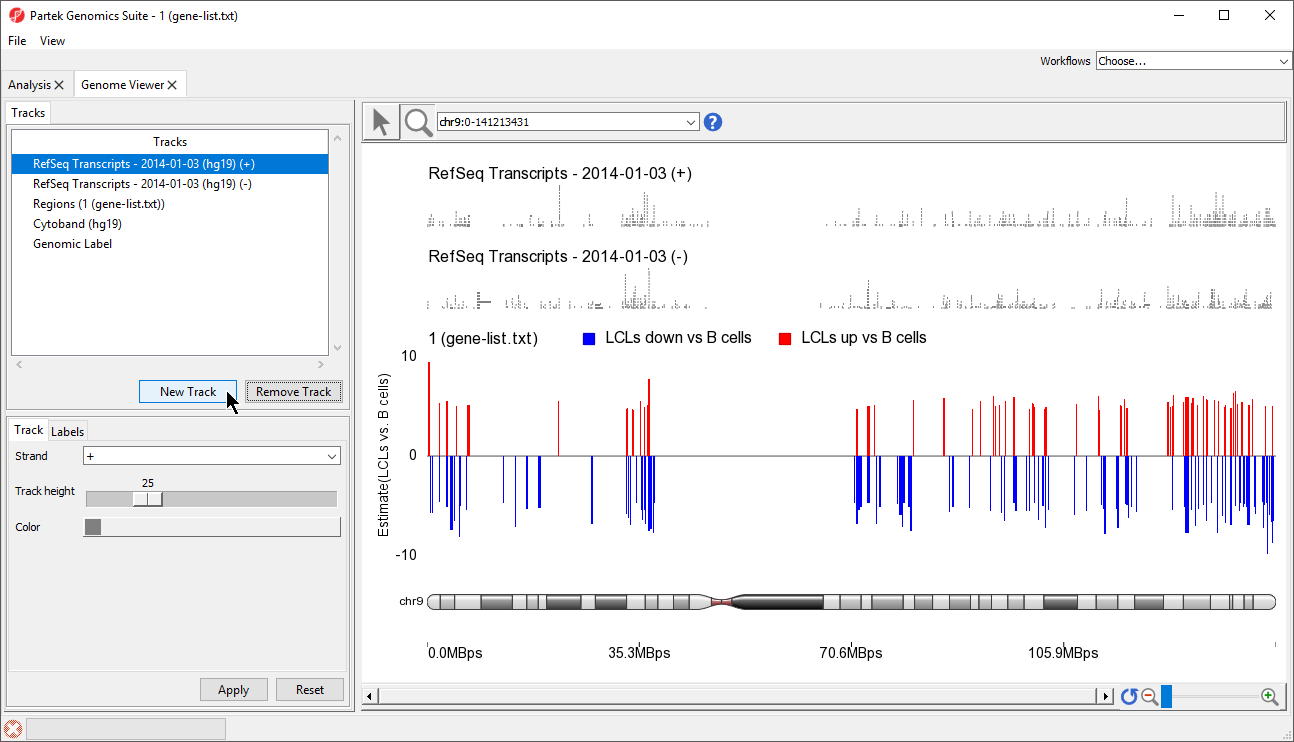
A new track will be created from the annotation file (Figure 9). If Separate Strands had been selected, there would be two tracks, one for each strand, like we see for the RefSeq Transcripts - 2014-01-03 (+) and (-) tracks (Figure 8).
|
|