...
- Click the Normalized counts node
- Click Differential analysis Statistics in the task menu
- Click GSA Differential analysis in the task menu (Figure 1)
| Numbered figure captions |
|---|
| SubtitleText | Invoking GSA from the task menuNavigating to the differential analysis options |
|---|
| AnchorName | Invoking GSADifferential analysis |
|---|
|
 Image Removed Image Removed
|
...
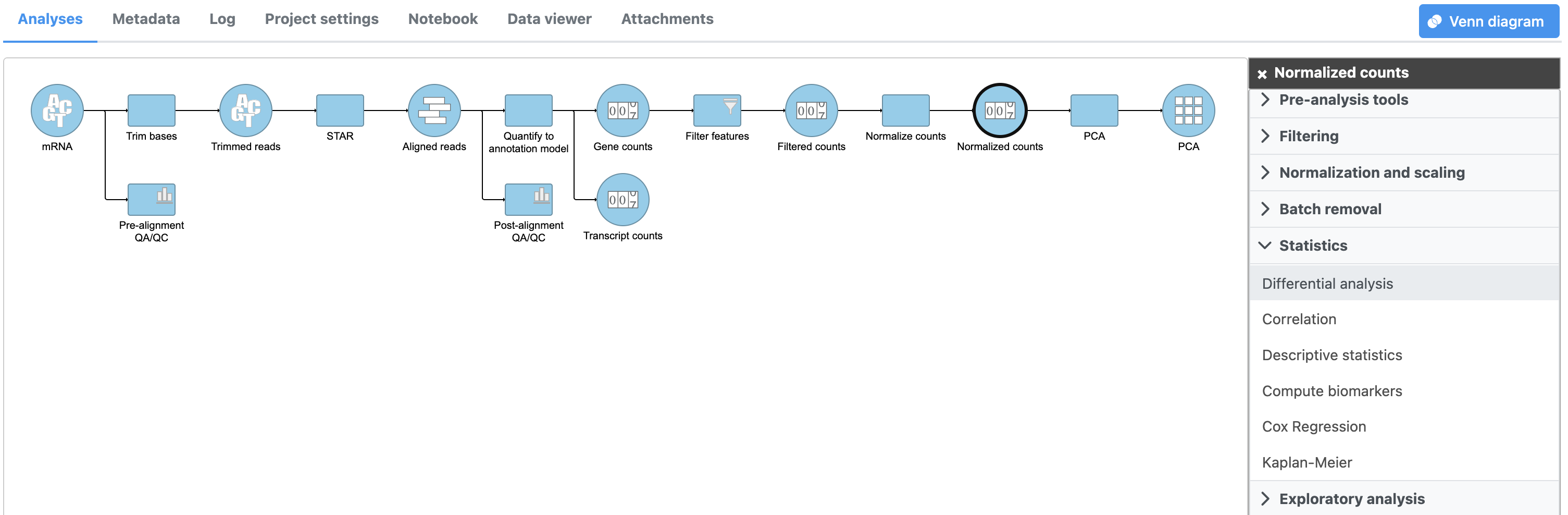 Image Added Image Added
|
Select the appropriate differential analysis method (Figure 2). Here, we only have one attribute, 5-AZA DoseIn this tutorial we are going to use DESeq2, but Partek® Flow® offers a number of alternatives. Hover the mouse over the  Image Added symbol for more information on each differential analysis method, or see our Differential Analysis user guide for a more in-depth look.
Image Added symbol for more information on each differential analysis method, or see our Differential Analysis user guide for a more in-depth look.
| Numbered figure captions |
|---|
| SubtitleText | Select the method for differential analysis from the options provided. |
|---|
| AnchorName | Method choice |
|---|
|
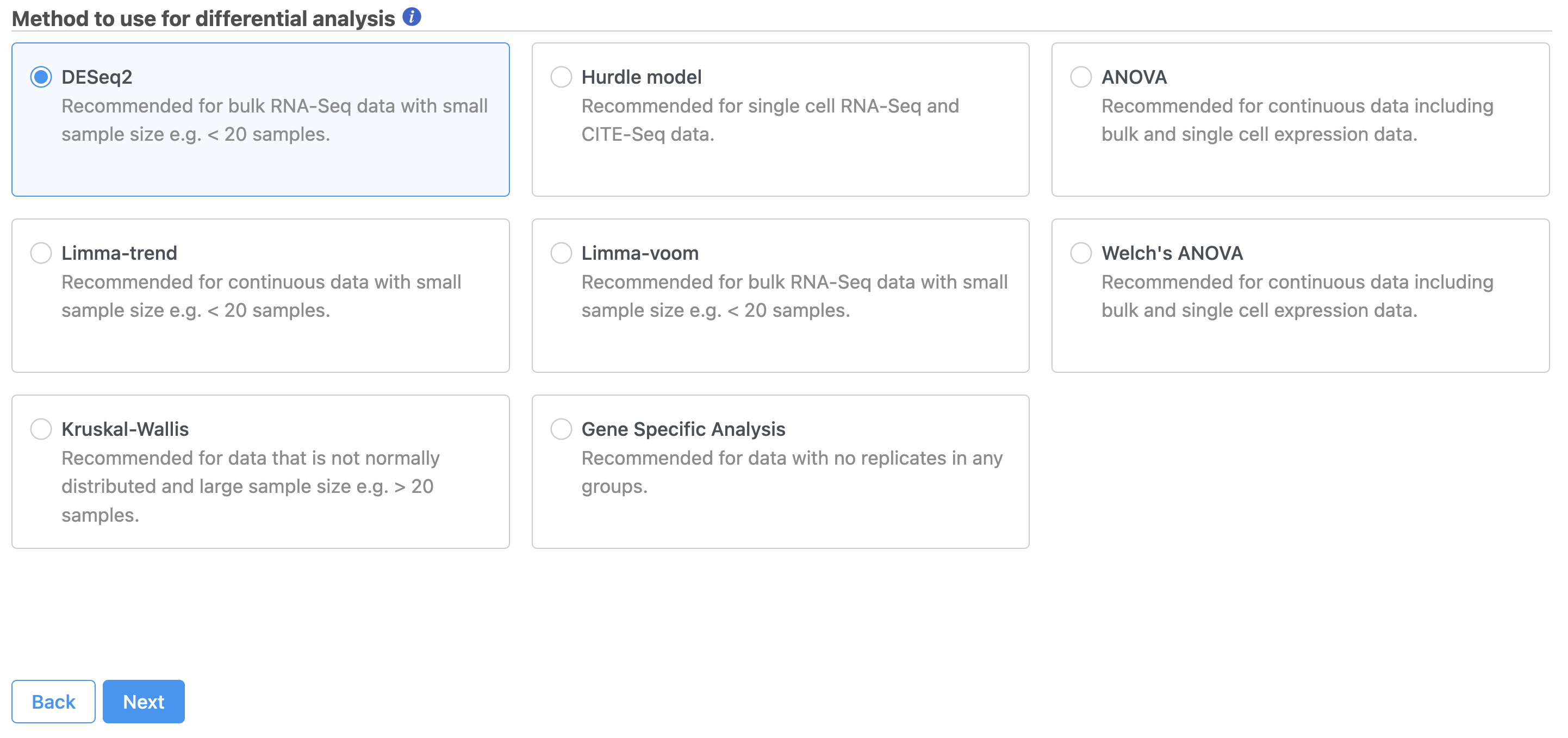 Image Added Image Added
|
- Check 5-AZA Dose and click Add factors to add the attribute to the statistical model.
| Numbered figure captions |
|---|
| SubtitleText | Selecting attributes to include |
|---|
| AnchorName | Include attributes |
|---|
|
 Image Removed Image Removed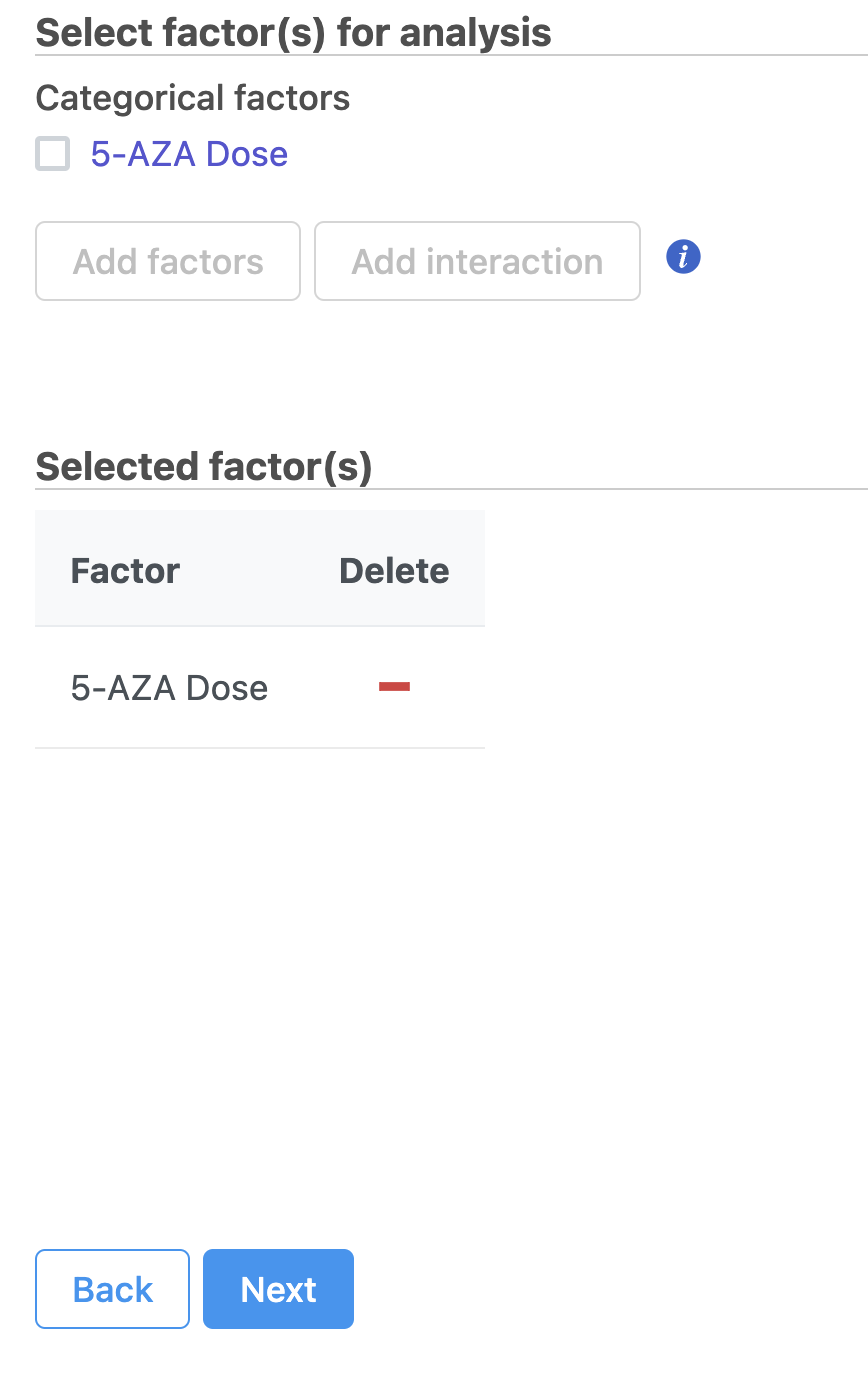 Image Added Image Added |
- Select Next to continue with 5-AZA Dose as the selected attribute
The Comparisons page will open (Figure 34).
| Numbered figure captions |
|---|
| SubtitleText | The Comparison selector allows multiple comparisons to be designed and added |
|---|
| AnchorName | Comparison selector |
|---|
|
 Image Removed Image Removed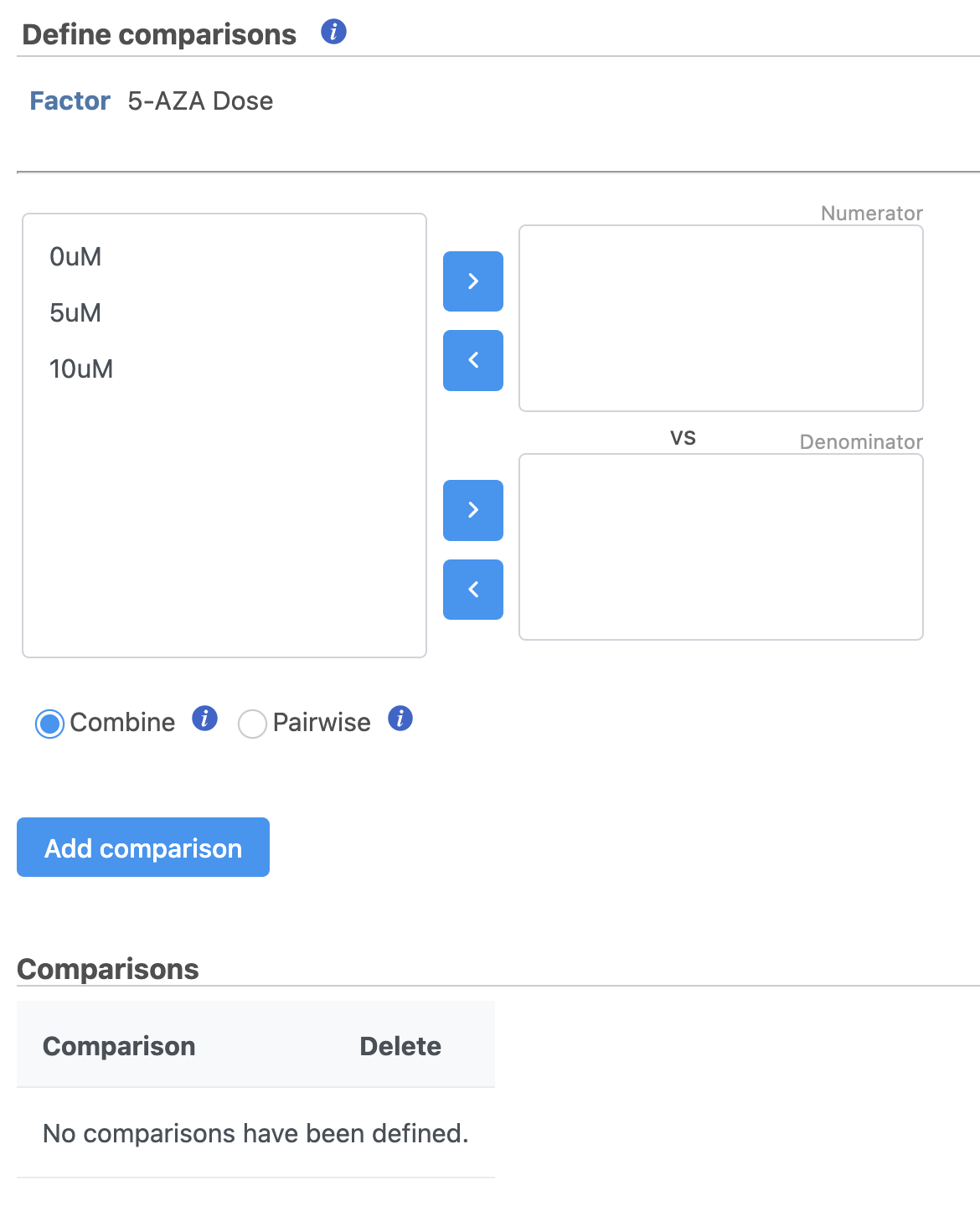 Image Added Image Added
|
It is easiest to think about comparisons as the questions we are asking. In this case, we want to know what are the differentially expressed genes between untreated and treated cells. We can ask this for each dose individually and for both collectively.
The upper box will be the numerator and the lower box will be the denominator in the comparison calculation so we will select the 0μM control in the lower box.
- Select Drag 5μM in to the upper box
- Select 0μM in Drag 0μM to the lower box
- Click Add comparison to add 5μM vs. 0μM to the comparison table (Figure 45)
| Numbered figure captions |
|---|
| SubtitleText | Designing a comparison to add |
|---|
| AnchorName | Adding comparisons |
|---|
|
 Image Removed Image Removed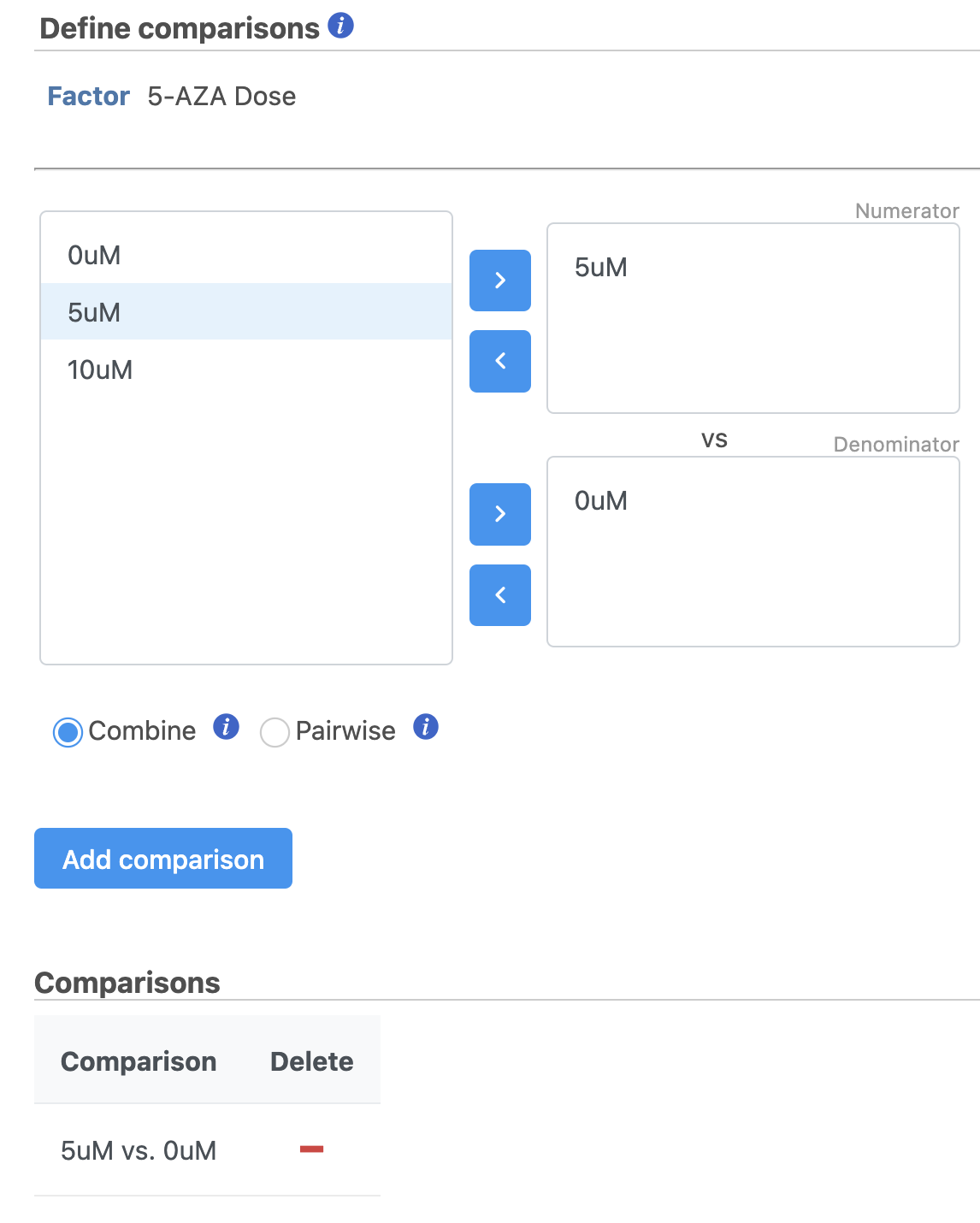 Image Added Image Added
|
- Repeat to create comparisons for 10μM vs. 0μM and 5μM:and 10μM,5μM vs. 0μM (Figure 56)
| Numbered figure captions |
|---|
| SubtitleText | Comparisons for 5uM vs. 0uM, 10uM vs. 0uM, and 5uM:10uM vs. 0uM have been added |
|---|
| AnchorName | Comparisons set up |
|---|
|
 Image Removed Image Removed
|
By default, the GSA will use a lognormal with shrinkage model for its analysis of variance of each gene. This is appropriate for most data sets and will tend to give accurate and reproducible results. To learn more about the advanced options available in the GSA task, please see the GSA user guide.
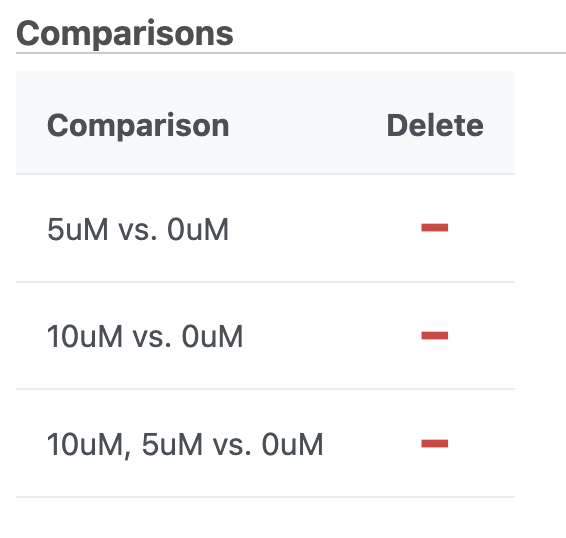 Image Added Image Added
|
- Click Finish to perform GSA DESeq2 as configured
A GSA DESeq2 task node and a GSA DESeq2 data node will be added to the pipeline (Figure 67).
| Numbered figure captions |
|---|
| SubtitleText | Gene analysis task node and Feature list data nodes |
|---|
| AnchorName | Gene analysis task node |
|---|
|
 Image Removed Image Removed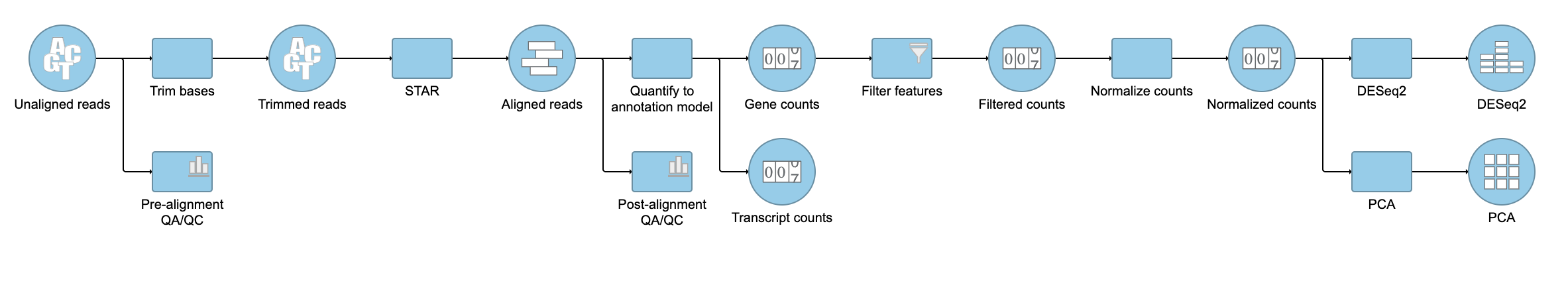 Image Added Image Added
|
...












