Page History
Next generation sequencing (NGS) data is notably huge in file size. Dealing with NGS data is not only time consuming but also puts constraints on hard disk space. This is especially true if analysis parameters need to be optimized. The Subsample FASTQ function is a very useful tool to get a subset of the raw data upon which optimization can be performed. The optimized parameters can then be saved and applied to the whole dataset
Subsample FASTQ is only available for unaligned reads of FASTQ format. To trigger this function, select the Unaligned Reads data node and select Subsample FASTQ from the Pre-alignment tools section on the menu. Then specify how many reads you want to keep for every nth reads. For example: if the user specifies to "Keep one read for every 10 reads" (Figure 1), this means that for every 10 reads, the program will keep only 1 read. This is equivalent to keeping 10% of the data.
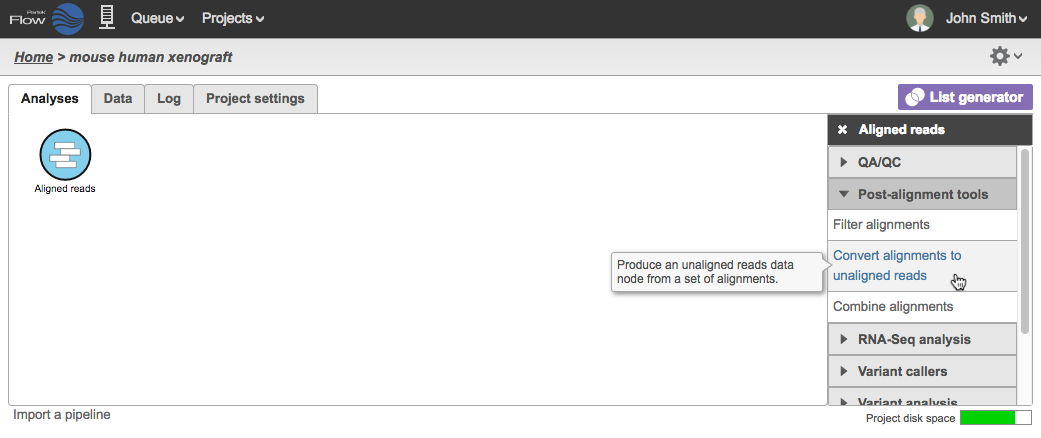
Aligned reads can be converted to unaligned reads in Partek Flow. The task is available under Post-alignment tools in the task menu when any Aligned reads data node is selected, which can be a result of an aligner in Partek Flow or data already aligned before import.
Generating unaligned reads from aligned data gives you the flexibility to remap the reads using either a different aligner, a different set of alignment parameters, or a different genome reference. This is particularly useful in analyzing sequences from xenograft models where the same set of reads can be aligned two different species. It may also be useful if the original unaligned FASTQ files are not as easily accessible to the user as the aligned BAM files.
To perform the task, select an Aligned reads data node and click Convert alignments to unaligned reads task in the task menu (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
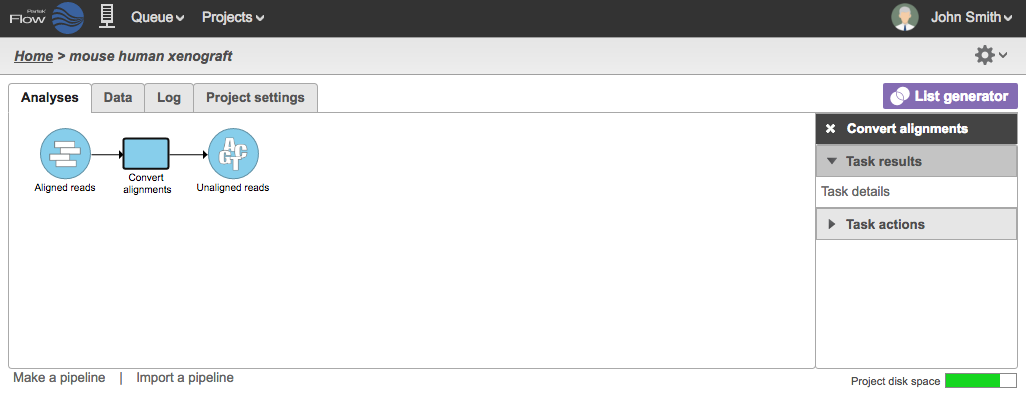
During the conversion, the BAM files are converted to FASTQ files and a new Unaligned reads data node will be generated (Figure 2) .
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
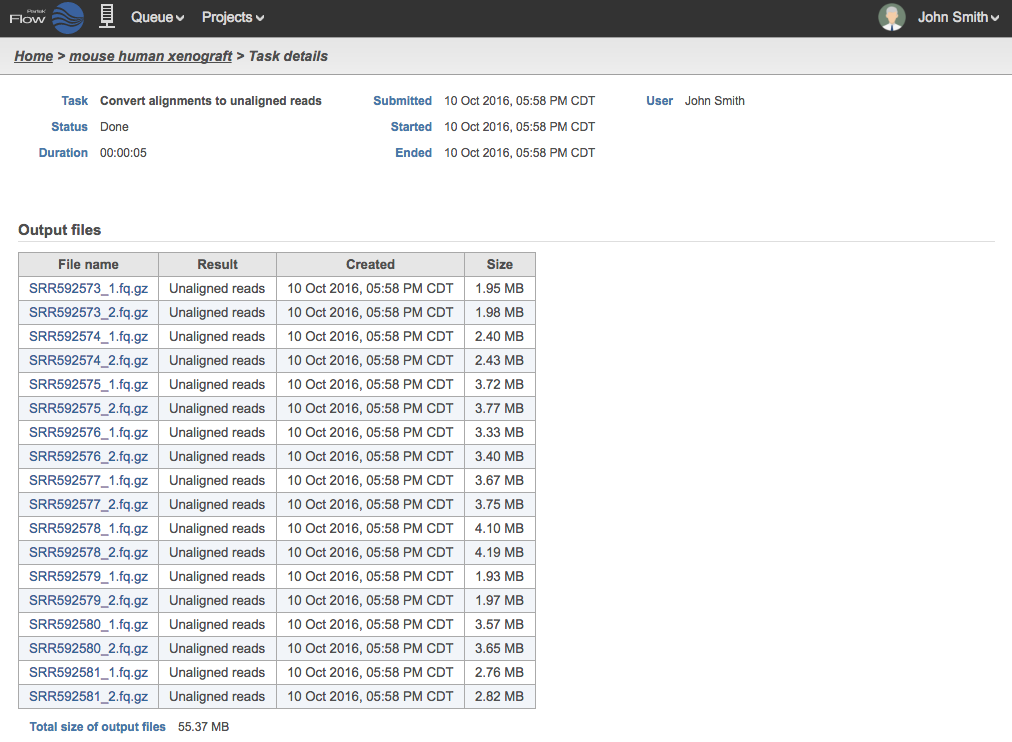
The filenames of the FASTQ files will be based on the sample names in the Data tab. The files generated are compressed with the extension *.fq.gz. For samples containing BAM files with paired end reads, two FASTQ files will be generated for each, and the files names will be appended with _1 and _2. An example in Figure 3 shows 18 .fq.gz output files that came from 9 BAM files.
| Numbered figure captions | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| |||||||
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...