...
With our reads trimmed, we now have high-quality reads for each sample. The next step is to align the reads to a reference genome. Alignment matches each of the short sequencing reads to a location in the reference genome.
- Select the Click Trimmed reads data node
- Select Click Aligners from in the task menu to display available aligners (Figure 1)
| Numbered figure captions |
|---|
| SubtitleText | A variety of different aligners are available in Partek Flow. In this tutorial we will use STAR aligner, a popular choice for aligning RNA-Seq data. |
|---|
| AnchorName | Invoking aligner |
|---|
|
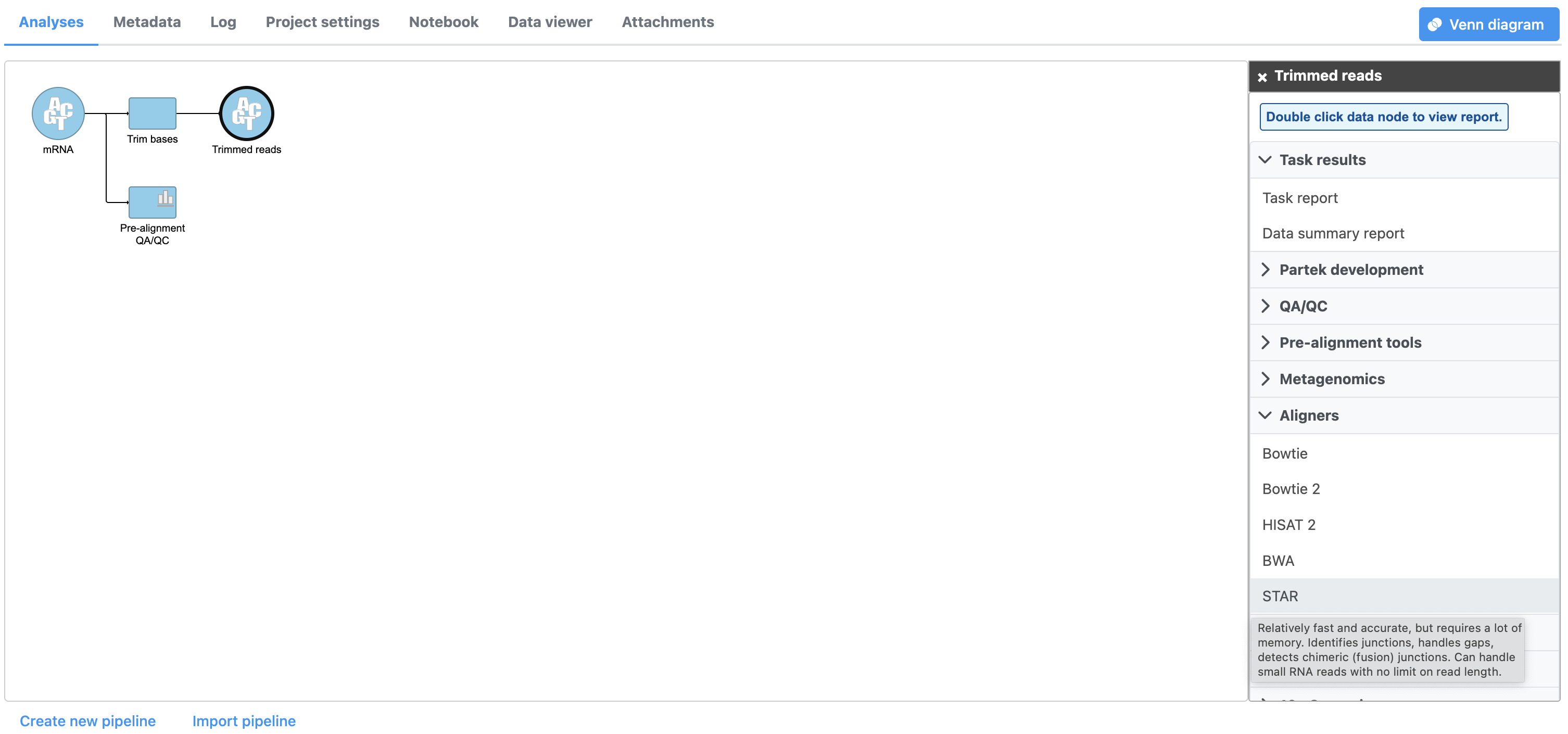 Image Added Image Added
|
Partek Flow offers a variety of different aligners. Mouse over any option for a short description. For this tutorial, we will use STAR, a fast and accurate aligner commonly used for RNA-Seq data. For more information about STAR and the other aligners, please consult the Aligners user guide.
- Select Click STAR from the task menu
The STAR aligner options allow us to select the genome build (assembly) and index. For this tutorial, our data set contains only using reads that map to chromosome 22 to minimize the time required for resource-intensive tasks, such as alignment.
- Select Homo sapiens (human) - hg19_chr22 from the Assembly drop-down menu
- Select Whole genome from the Aligner index drop-down menu
- Select Finish
- Click Finish to run with hg19 selected for Assembly and Whole genome for the Aligner index (Figure 2)
| Numbered figure captions |
|---|
| SubtitleText | Configuring the STAR aligner |
|---|
| AnchorName | Configuring STAR aligner |
|---|
|
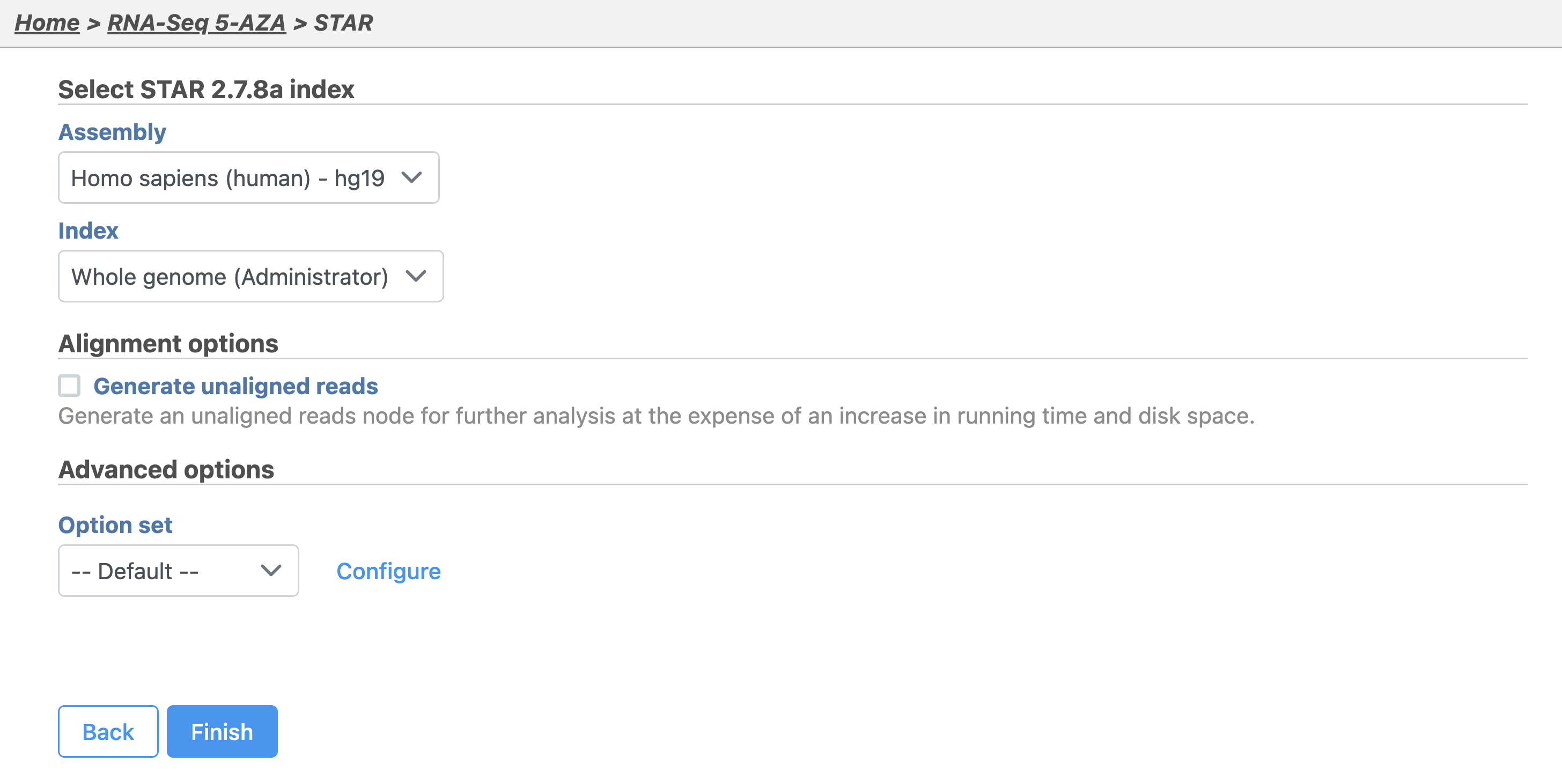 Image Added Image Added
|
Alignment is a resource-intensive task and may take a while around 20 minutes to complete, even when mapping only to reads from a single chromosome 22.
- Mouse over the Align reads task node to view the estimated end time and progress percentage (Figure)
As we can see, the Align reads task will generate . Task and data nodes that have been queued, but not completed, are shown in a ligher color than completed tasks (Figure 3).
| Numbered figure captions |
|---|
| SubtitleText | Queued tasks and their output data nodes are shown in a lighter color |
|---|
| AnchorName | Queued tasks |
|---|
|
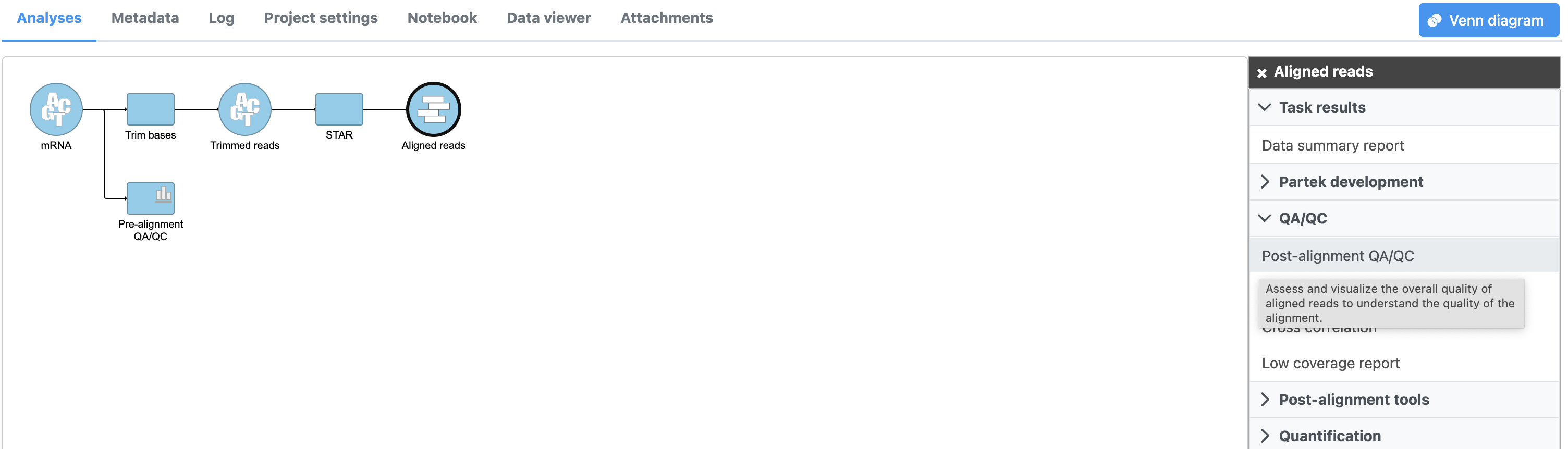 Image Added Image Added
|
The Align reads task generates an Aligned reads data node once complete. If you would like, you can return to Partek Flow when You can wait for the alignment task has completed, to finish or you can continue building the pipeline while the results of alignment are pending. Additional ; additional tasks can be added to the pipeline and queued before the current task has completed.
|
...


