Page History
...
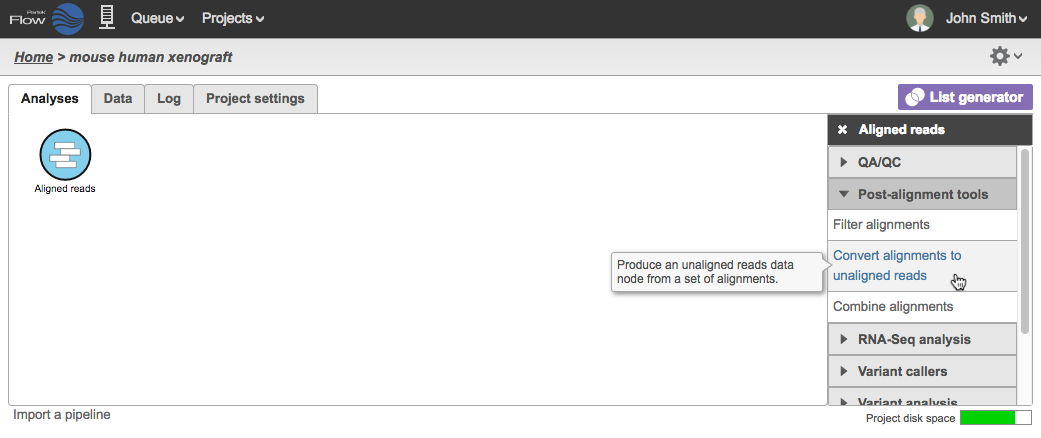
To perform the task, select an aligned reads data node and click Convert alignments to unaligned reads task in the task menu (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
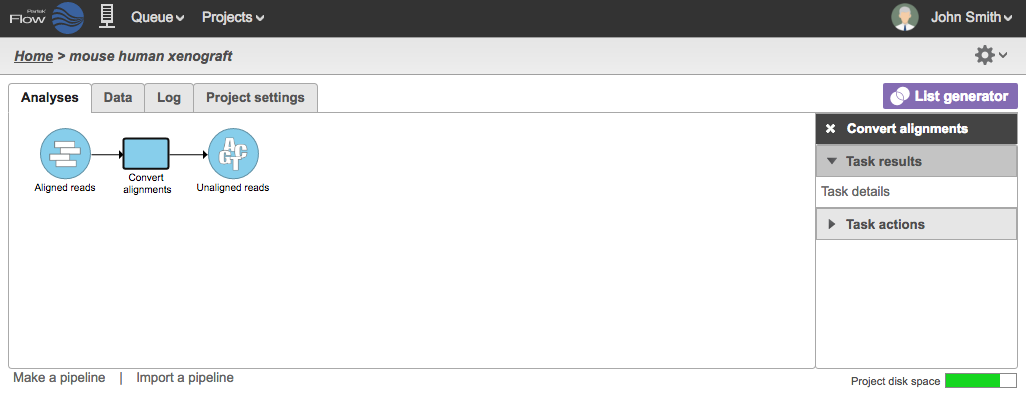
A new data node will be generated containing FASTQ files (Figure 2) . Note that the files generated are compressed and the filenames are *.fastqfq.gz.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
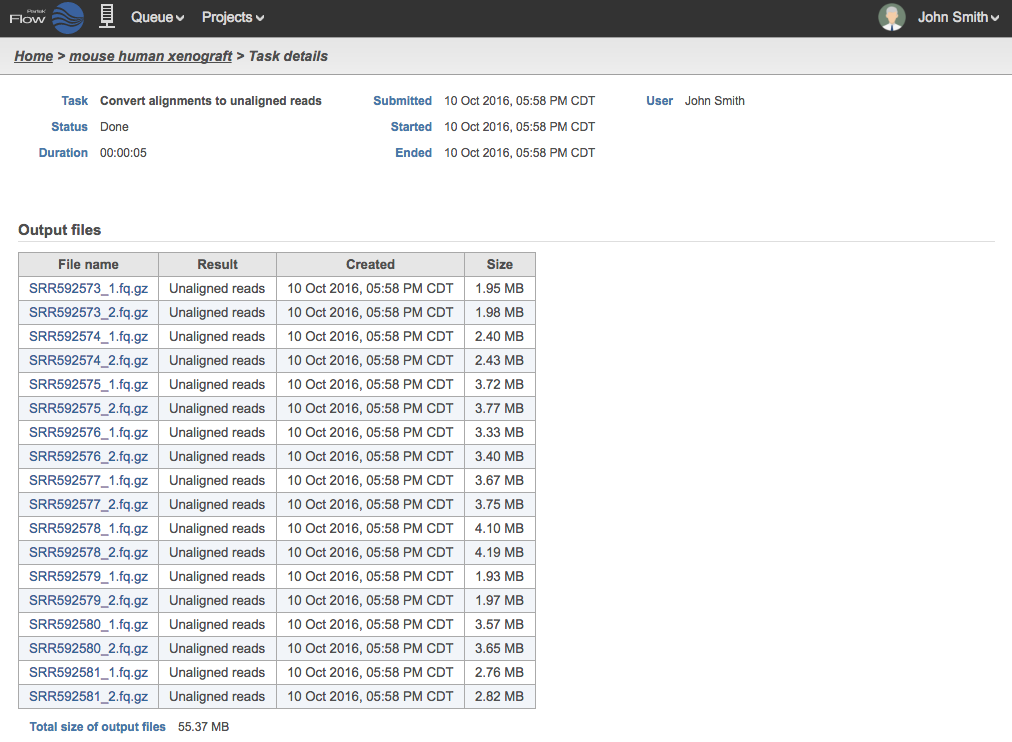
During the conversion, the BAM files are converted to FASTQ files. If the BAM The name of the BAM files will be based on the sample names in the Data tab. If the BAM file contains paired end reads, two FASTQ files will be generated for each BAM, and the files names will contain _1 and _2. An example in Figure 3 shows 18 fastqFQ.gz GZ files from 9 .bam BAM files.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|
...
Overview
Content Tools