Page History
...
Exploratory data analysis
Principal Components Analysis (PCA) is an excellent method to visualize similarities and differences between the samples in a data set. PCA can be invoked through a workflow, by selecting () from the main command bar, or by selecting Scatter Plot from the View section of the main toolbar. We will use a workflow.
Select the Affy_miR_BrainHeart_intensities spreadsheet
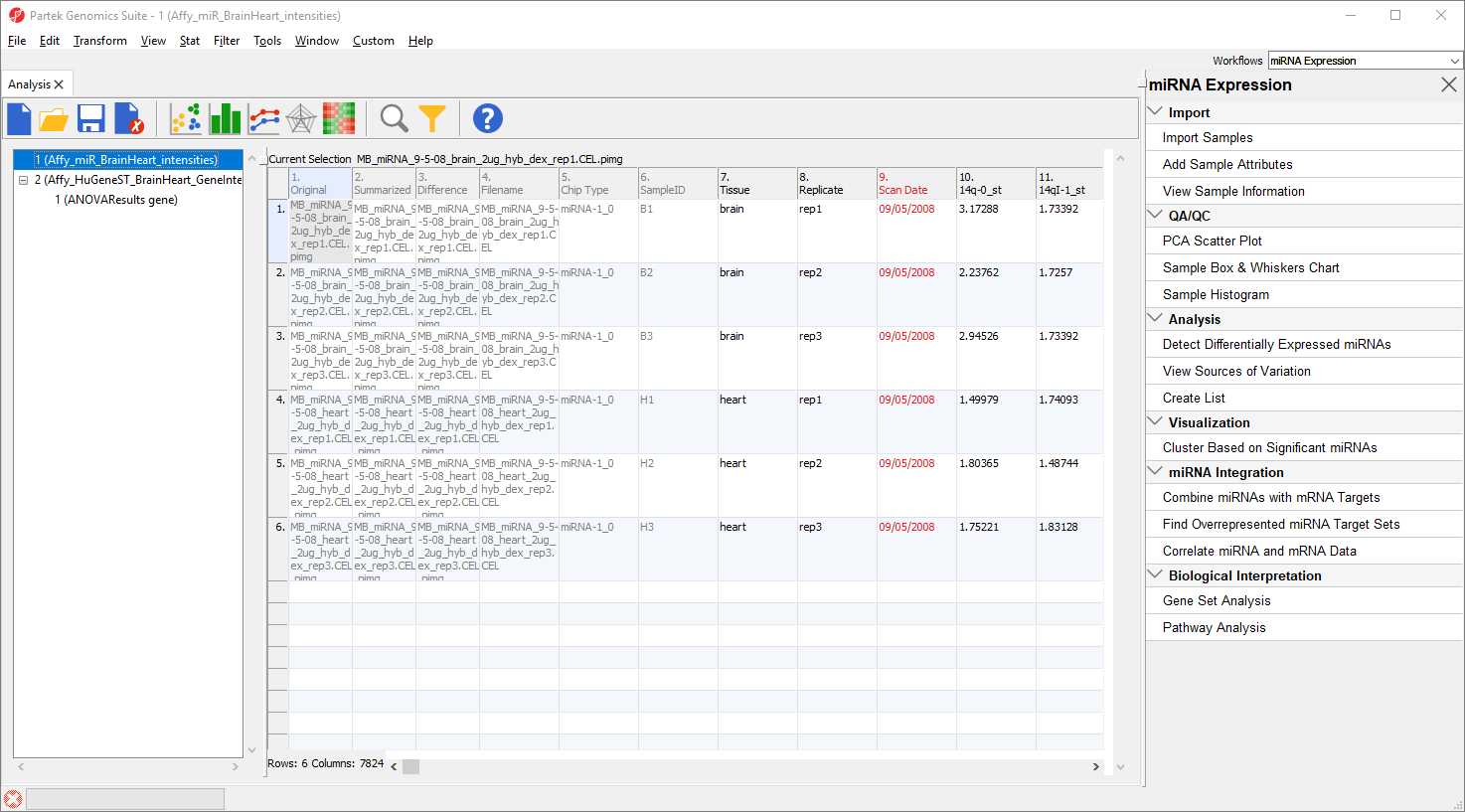
This is the probe intensities spreadsheet for the miRNA expression data (Figure 2). Each row is a sample; columns 7 to 9 give attribute information about each sample including tissue, replicate number, and scan date, while columns 10 on give prove intensities values.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select PCA Scatter Plot from the QA/QC section of the workflow
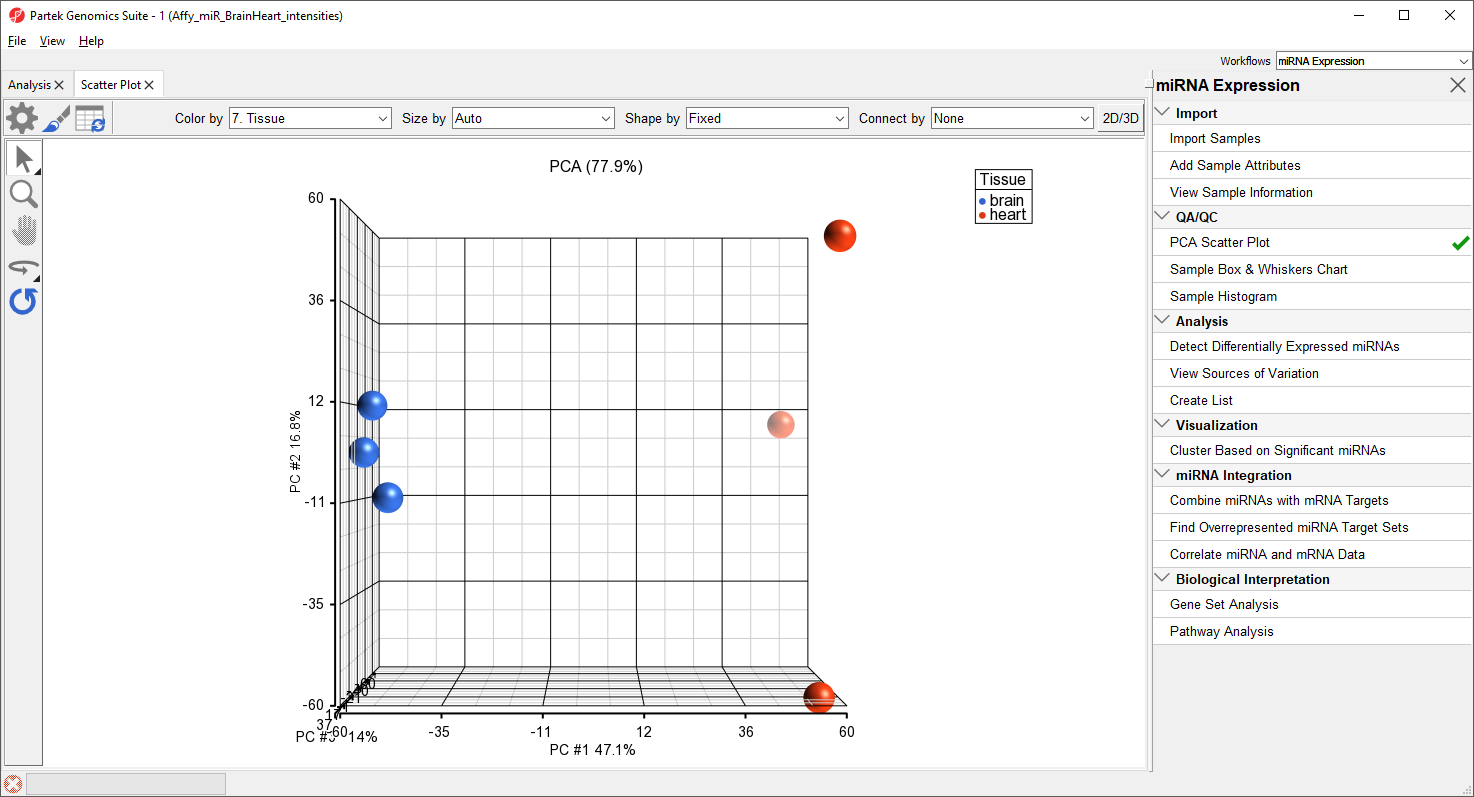
A new tab will open showing a PCA scatter plot (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
In this PCA scatter plot, each point represents a sample in the spreadsheet. Points that are close together in the plot are more similar, while points that are far apart in the plot are more dissimilar.
To better view the data, we can rotate the plot.
- Select () to activate Rotate Mode
- Click and drag to rotate the plot
Rotating the plot allows us to look for outliers in the data on each of the three principal components (PC1-3). The percentage of the total variation explained by each PC is listed by its axis label. The chart label shows the sum percentage of the total variation explained by the displayed PCs.
Here, we can see that the brain and heart samples are well separated across PC1, which is expected.
For more information about customizing the plot, please see Exploring the data set with PCA from the Gene Expression with Batch Effect tutorial.
Detecting differentially expressed miRNAs
Next, we will identify miRNAs that are differentially expressed between brain and heart tissues.
- Selectthe Analysis tab
- Select the Affy_miR_BrainHeart_intensities spreadsheet
- Selelct Detect Differentially Expressed miRNAs from the Analysis section of the workflow
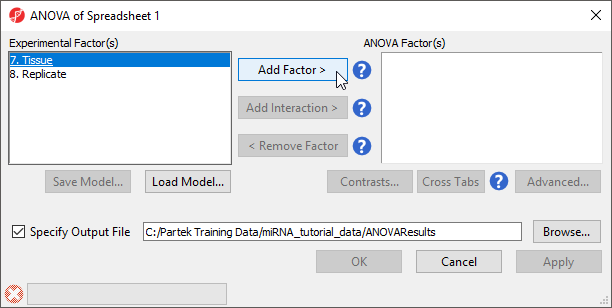
The ANOVA dialog (Figure 4) allows us to configure the comparisons we want to make between samples and groups within the data set.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Tissue from the Experimental Factor(s) panel
- Select Add Factor > to move Tissue to the ANOVA Factor(s) panel
The Contrasts... button will now be available to select.
- Select Contrasts...
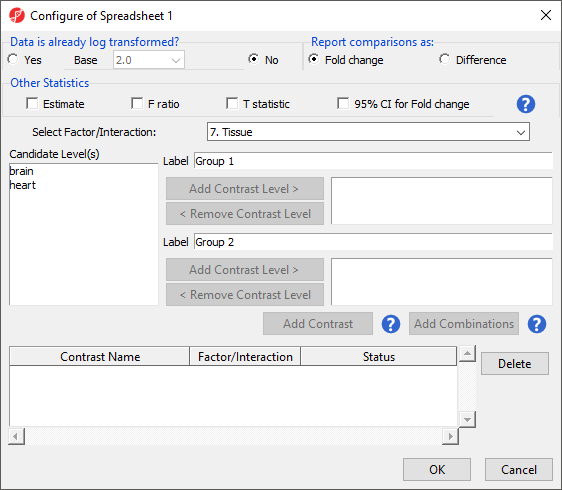
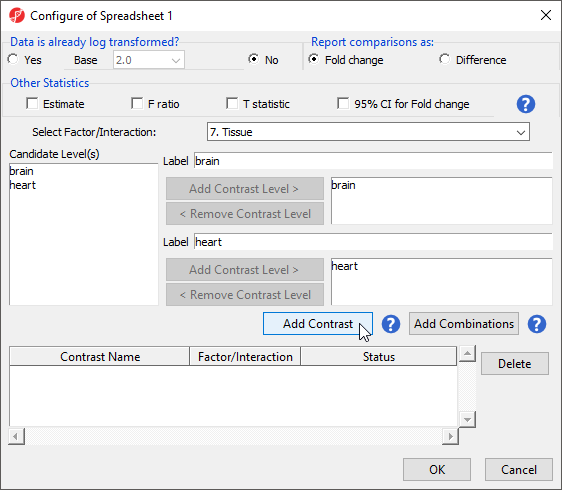
The Configure ANOVA dialog (Figure 5) is used to set up contrasts. Contrasts are the comparisons between groups and are where experimental questions can be asked. In this study, we are asking what miRNAs are differentially expressed between heart and brain tissue.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Yes forData is already log transformed?
- Select Fold change for Report comparisons as
- Select 7. Tissue from the Select Factor/Interaction drop-down menu
- Select brain from the left panel
- Select Add Contrast Level > to move brain to the upper group - initially Group 1
- Select heart from the left panel
- Select Add Contrast Level > to move heart to the lower group - initially Group 2
This contrast (Figure 6) will compare expression of miRNAs in brain samples to expression in heart samples with brain as the numerator and heart as the denominator for fold-change calculations.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Add Contrast
- Select OK
The Contrasts... button should now read Contrasts Included.
- Select OK to run the ANOVA as configured
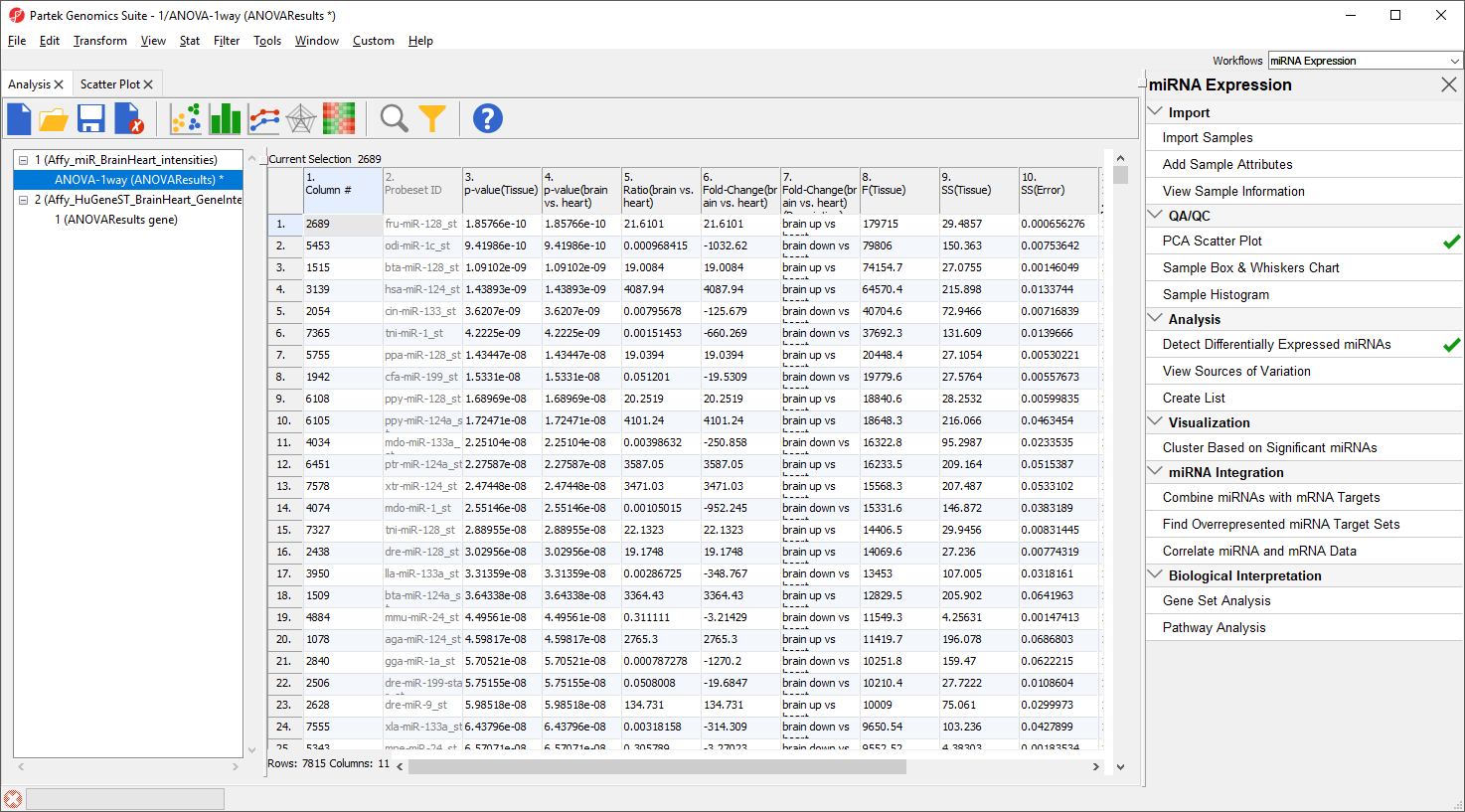
An ANOVA Results sheet, ANOVAResults, will be created as a child spreadsheet of Affy_miR_BrainHeart_intensities (Figure 7). In this spreadsheet, each row represents a probe set and the columns represent the computation results for that probe set. Although not synonymous, probe set and gene will be treated as synonyms in this tutorial for convenience. By default, the genes are sorted in ascending order by the p-value of the first categorical factor, which, in this case, is Tissue. This means the most significant differentially expressed miRNAs between the brain and heart (up-regulated and donw-regulated) are at the top of the spreadsheet.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
You may explore what is known about any listed microRNA using external databases TargetScan, miRBase, microRNA.org, or miR2Disease, by right-clicking a row header, selecting Find miRNA in... and choosing one of the external databases. This will open a web page in your default web browser and requires your computer be connected to the internet.
For more information about AVOVA in Partek Genomics Suite, see Identifying differentially expressed genes using ANOVA.
Creating a list of miRNAs of interest
The ANOVA results spreadsheet includes every miRNA on the array for a total of 7815 miRNAs. However, many of these miRNAs are not significantly differentially expressed between brain and heart and, thus, are not of interest. Next, we will create a filtered list of significantly differentially expressed miRNAs.
- Select the ANOVAResults spreadsheet
- Select Create List from the Analysis section of the workflow
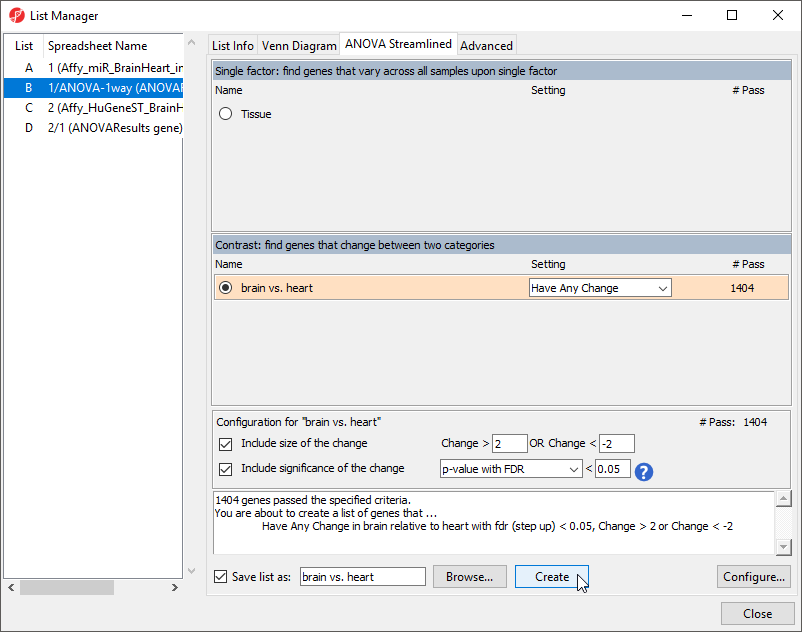
The List Manager dialog will open (Figure 8).
- Select brain vs. heart under Contrast: find genes that change between two categories
By default, the fold-change and significance thresholds are set to > 2, < -2 and p-value with FDR < 0.05. These defaults are appropriate for this tutorial so we will leave them in place.
- Select Create to create a new list, brain vs. heart containing only the 1404 miRNAs that pass the criteria
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A new spreadsheet, brain vs. heart will be created as a child spreadsheet of Affy_miR_BrainHeart.
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|