This tutorial will illustrate:
Note: the workflow described below is enabled in Partek Genomics Suite version 7.0 software. Please fill out the form on Our support page to request this version or use the Help > Check for Updates command to check whether you have the latest released version. The screenshots shown within this tutorial may vary across platforms and across different versions of Partek Genomics Suite.
Introduction to Survival Analysis
Survival analysis is a branch of statistics that deals with modeling of time-to-event. In the context of “survival,” the most common event studied is death; however, any other important biological event could be analyzed in a similar fashion (e.g., spreading of the primary tumor or occurrence/relapse of disease). The significant event should be well-defined and occur at a specific time. As the primary outcome event is typically unfavorable (e.g., death, metastasis, relapse, etc.), the event is called a “hazard.” Survival analysis tries to answer questions such as: What is the proportion of a population who will survive past a certain time (i.e., what is the 5-year survival rate)? What is the rate at which the event occurs? Do particular characteristics have an impact on survival rates (e.g., are certain genes associated with survival)? Is the 5-year survival rate improved in patients treated by a new drug?
An important feature of survival analysis is the presence of “censored” data. Censored data refers to subjects that have not experienced the event being studied. For example, medical studies often focus on survival of patients after treatment so the survival times are recorded during the study period. At the end of the study period, some patients are dead, some patients are alive, and the status of some patients is unknown because they dropped out of the study. Censored data refers to the latter two groups. The patients who survived until the end of the study or those who dropped out of the study have not experienced the study event "death" and are listed as "censored".
Tutorial Data Set
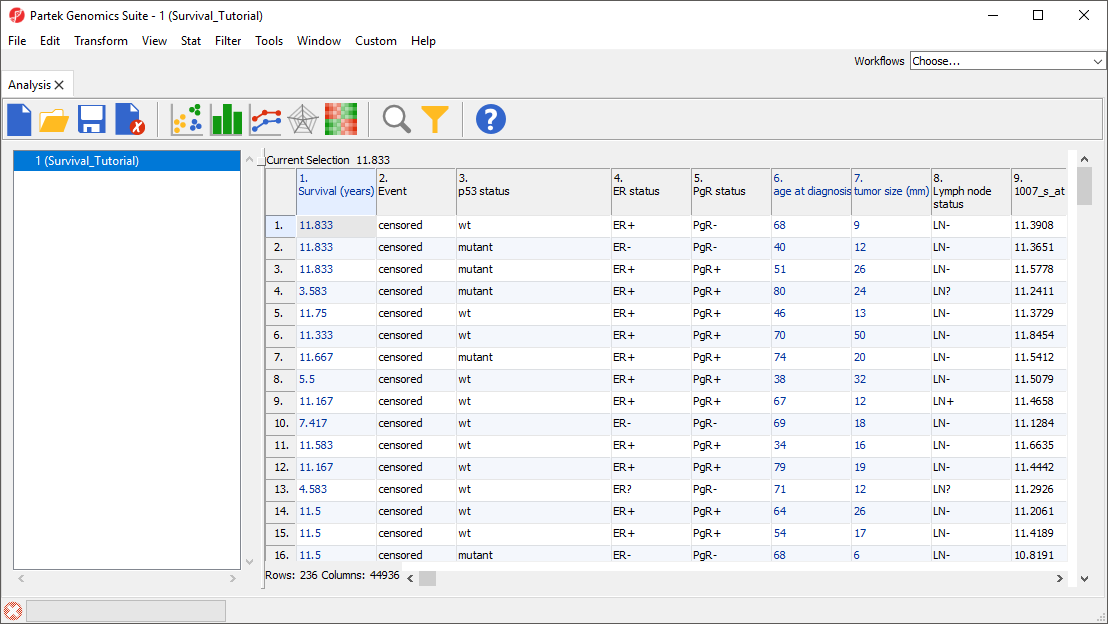
The tutorial data set (236 samples) is a subset of fresh-frozen breast tumor specimens from a population-based cohort of 315 women with breast cancer. The clinicopathological characteristics accompanying each tumor include p53 status (mutant or wild-type), estrogen receptor (ER) status, progesterone receptor (PgR) status, lymph node status, tumor size, and patient age. Gene expression was assessed on Affymetrix® U133A and U133B arrays (Miller LD et al., GSE3494). Please note that Affymetrix data have been chosen for the illustration purposes only, and that the same functionality can be used to analyze any data set. The raw data files (.CEL) have already been imported into PGS; samples with no survival time data, as well as sample attributes irrelevant for the survival analysis, were removed, and the final spreadsheet was saved in Partek Genomics Suite (Survival_Tutorial.fmt and Survival_Tutorial.txt). To go through the tutorial, download the tutorial data set, unzip the downloaded folder and save it in an easily accessible location on your computer.
After saving the unzipped file, you can open it in Partek Genomics Suite.
- Select File from the main toolbar
- Select Open...
- Browse to the folder containing the tutorial data set and select the file Survival_Tutorial.fmt
The data spreadsheet will open (Figure 1). Each row represents a tumor sample from a breast cancer patient. Sample attributes are listed in columns 1-8, while columns 9+ are intensity values for the probe sets listed in the column headers.
References
Miller LD, Smeds J, George J, Vega VB et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. PNAS, 2005; 102(38): 13550-5.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
36 | rates |