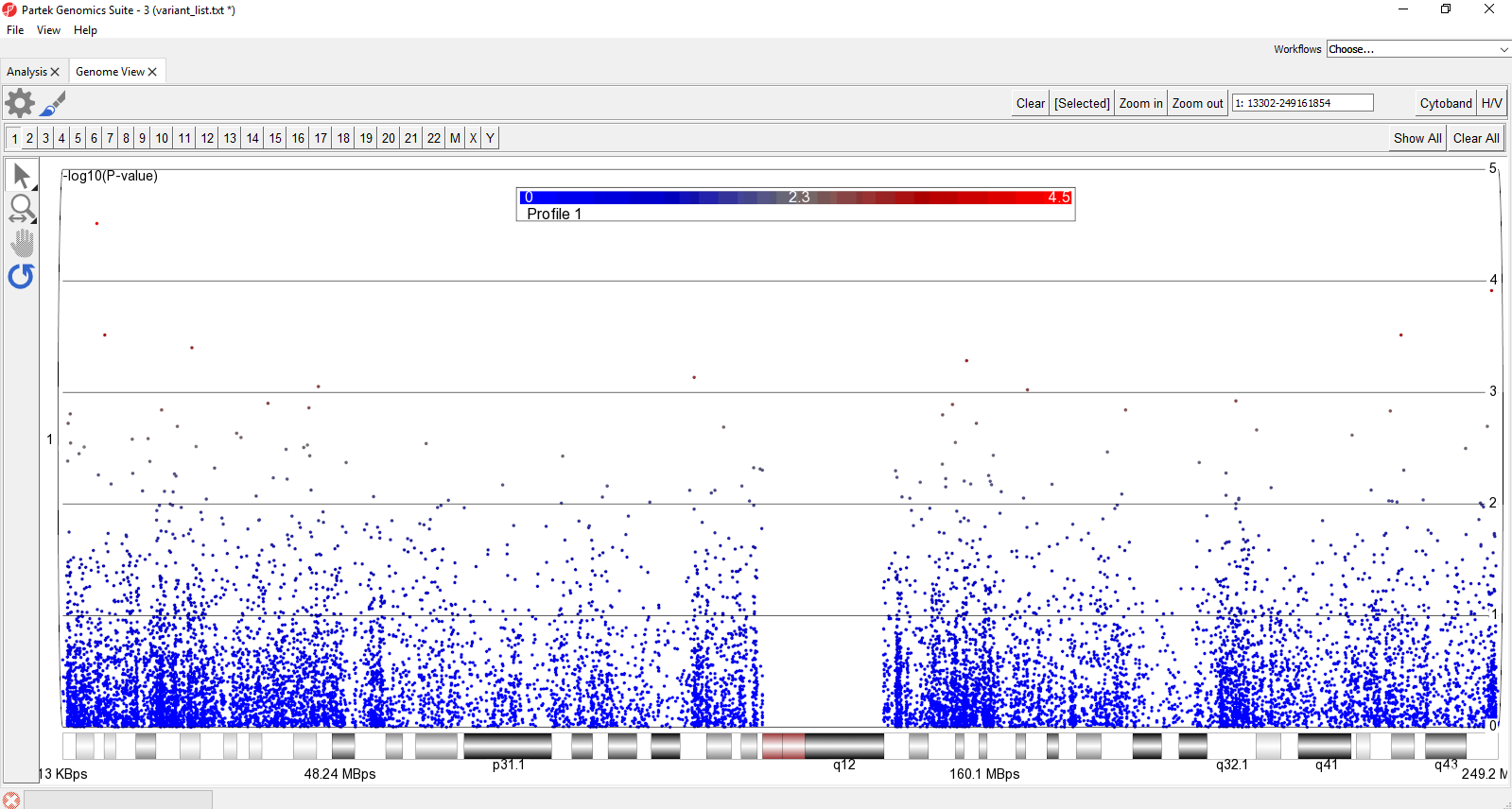
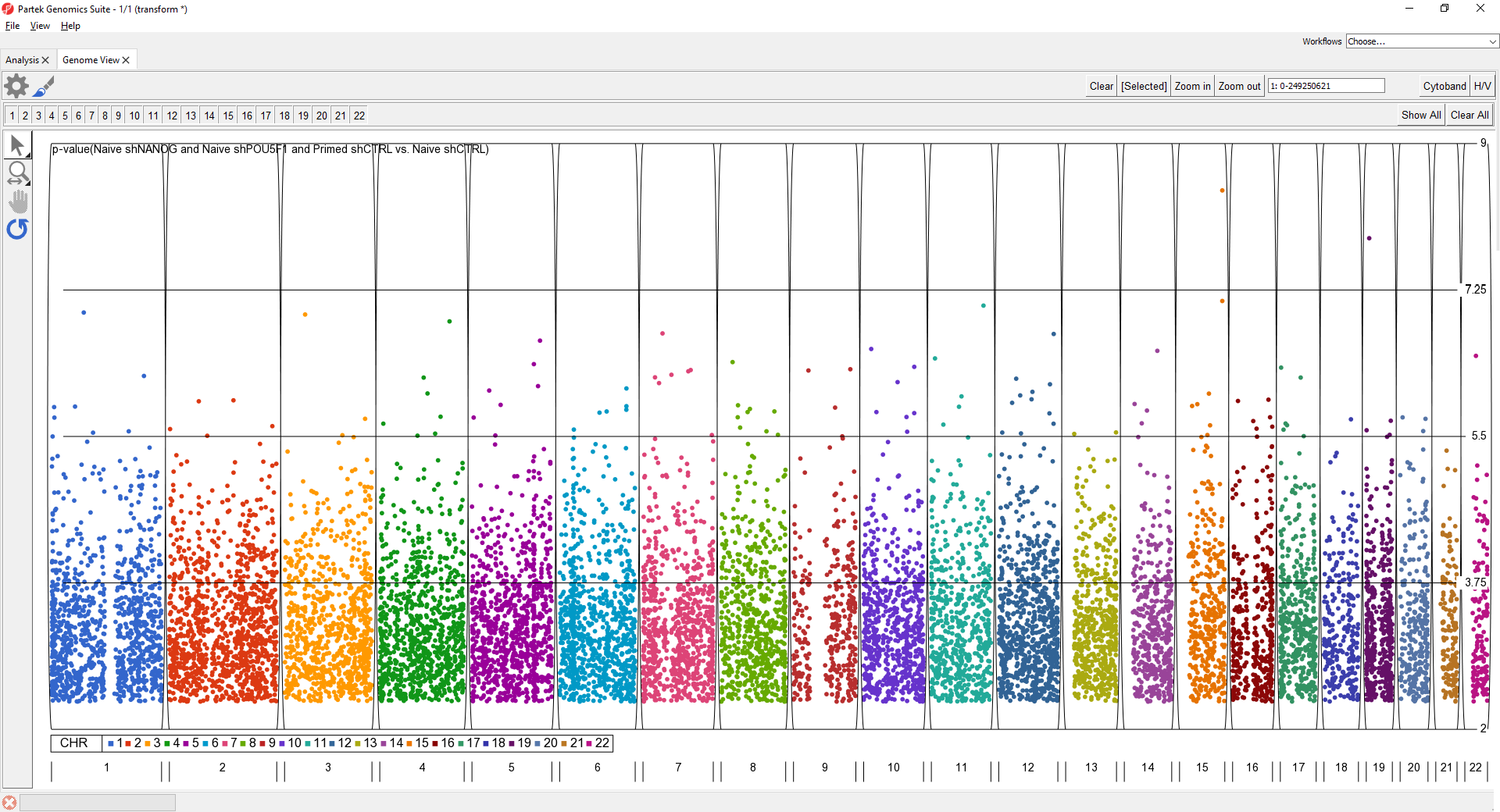
The Manhattan plot is a common way to visualize p-values or log-odds ratios for GWAS studies across genomic coordinates.
The starting point for a Manhattan plot is a spreadsheet with SNPs on rows and p-values or log-odds ratios in a column. If beginning with p-values, you will need to convert the p-values to -log10(p-value).
- Select the column with p-values
- Select Transform form the main toolbar
- Select Normalization & Scaling
- Select On Columns...
- In the Normalization tab, set Base of the Log(x + offset) to 10
- Select OK
- Go to Transform > Normalization & Scaling > On Columns... again
- Select the Add/Mul/Sub/Div tab
- Set Multiply by Constant to -1
- Select OK
The column now contains -log10(p-value).
We can now invoke the initial plot.
- Select View from the main toolbar
- Select Genome View
The Genome View tab will open. This plot will need to be configured.
- Select () from the plot command bar
- Select the Profiles tab
- Remove any unwanted profiles
- Select Add profile
- Select Column
- Select the column with the -log10(p-value) or logs-odds ratio values from the drop-down menu
- Select Value for Color by
- Select point from the Style drop-down menu
- Select OK to add the profile
- Select OK to close the Configure Plot Properties dialog
The plot will now show a Manhattan plot (Figure 1).
- Select Show All in the upper-right hand corner of the plot
This displays all chromosomes vertically. We can display them horizontally for a better view.
- Select to open the Configure Plot dialog
- Select Genome in line for Layout
- Select OK
To further improve the genome-wide view, we can remove the cytoband, remove the genomic position label, color points by chromosome, and increase point size.
- Select Cytoband in the upper right-hand corner
- Select
- Deselect Show Base Pair Labels
- Select Profiles
- Select Configure
- Set Color By to a column with chromosome for each SNP/loci as a category
- Set Shape Size to 5.0
- Select OK to close the Configure Profile dialog
- Select OK to apply changes
The plot will appear as shown (Figure 2).
For details on Genome View see Chapter 6: The Pattern Visualization System in the Partek User's Manual.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
34 | rates |